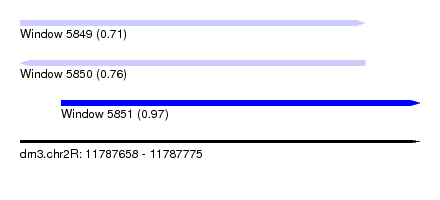
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,787,658 – 11,787,775 |
| Length | 117 |
| Max. P | 0.974265 |

| Location | 11,787,658 – 11,787,759 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 67.90 |
| Shannon entropy | 0.57618 |
| G+C content | 0.53008 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -15.56 |
| Energy contribution | -15.26 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
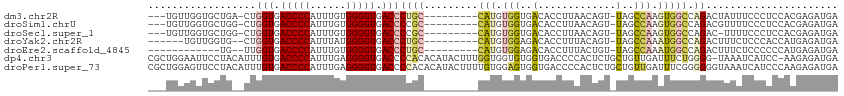
>dm3.chr2R 11787658 101 + 21146708 ---UGUUGGUGCUGA-CUGGUGACCCCAUUUGUGGGGUGACCCUGC---------CAUGUGGUGACACCUUAACAGU-UAGCCAAGUGGCCAGACUAUUUCCCUCCACGAGAUGA ---..((((((((((-(((((.((((((....)))))).)).....---------...(((....))).....))))-))))......)))))..((((((.......)))))). ( -34.80, z-score = -1.43, R) >droSim1.chrU 12444360 101 + 15797150 ---UGUUGGUGCUGG-CUGGUGACCCCAUUUGUGGGGUGACCCCGC---------CAUGUGGUGACACCUUAACAGU-UAGCCAAGUGGCCAGACGUUUUCCCUCCACGAGAUGA ---..((((((((((-(.(((.((((((....)))))).)))..))---------)..(((....)))......)))-..)))))((((...((.....))...))))....... ( -36.70, z-score = -1.21, R) >droSec1.super_1 9283535 100 + 14215200 ---UGUUGGUGCUGG-CUGGUGACCCCAUUUGUGGGGUGACCCCGC---------CAUGUGGUGACACCUUAACAGU-UAGCCAAGUGGCCAGAC-UUUUCCCUCCACGAGAUGA ---..((((((((((-(.(((.((((((....)))))).)))..))---------)..(((....)))......)))-..)))))((((...((.-...))...))))....... ( -36.70, z-score = -1.36, R) >droYak2.chr2R 3944288 97 - 21139217 ------UGUUGGUG--CUGGUGACCCCAUUUAUGGGGUGACCCUGC---------CAUGUGGAGACACCUUUACAGU-UAGCCAAAUGGCCAGACUUUCUCCCACCAUGAGAUGA ------...(((((--..(((.((((((....)))))).)))((((---------(((.(((.(((.........))-)..))).)))).))).........)))))........ ( -33.30, z-score = -1.80, R) >droEre2.scaffold_4845 8518659 91 - 22589142 ------------UG--UUGGUGACCCCAUUUGUGGGGUGACCCUGC---------CAUGUGGAGACACCUUUACUGU-UAGCCAAAUGGCCAGACUUUCUCCCCCCAUGAGAUGA ------------..--..(((.((((((....)))))).)))((((---------(((.(((.((((.......)))-)..))).)))).)))....((((.......))))... ( -30.90, z-score = -2.32, R) >dp4.chr3 17847152 113 + 19779522 CGCUGGAAUUCCUACAUUUGUGACCCCAUUUGAGGGGUGACCCCACACAUACUUUGGUGGUGUGGUGACCCCACUCUGCUGUUGAUUUCUGGGG-UAAAUCAUCC-AAGAGAUGA .(..((....))..)....((.(((((......))))).)).((((((.(((....)))))))))..((((((.....(....).....)))))-)...(((((.-....))))) ( -35.90, z-score = -0.56, R) >droPer1.super_73 63954 115 - 267513 CGCUGGAGUUCCUACAUUUGUGACCCCAUUUGAGGGGUGACCCCACACAUACUUUUGUGGAGUGGUGACCCCACUCUGCUGUUGAUUUCGGGGGGUAAAUCAUCCCAAGAGAUGA .((.(((((.....((..((......))..)).(((((.((((..((((......))))..).))).)))))))))))).....(((((..(((((.....)))))..))))).. ( -40.50, z-score = -1.12, R) >consensus ___UGUUGGUGCUG__CUGGUGACCCCAUUUGUGGGGUGACCCCGC_________CAUGUGGUGACACCUUAACAGU_UAGCCAAGUGGCCAGACUUUUUCCCCCCACGAGAUGA ..................(((.(((((......))))).))).................(((..(.((.......)).)..))).((((...((.....))...))))....... (-15.56 = -15.26 + -0.30)
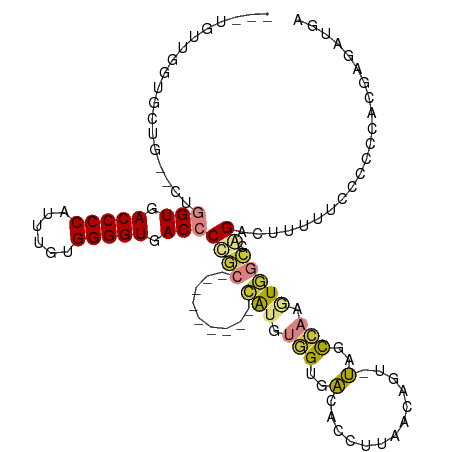
| Location | 11,787,658 – 11,787,759 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 67.90 |
| Shannon entropy | 0.57618 |
| G+C content | 0.53008 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -15.92 |
| Energy contribution | -17.74 |
| Covariance contribution | 1.82 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.755715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
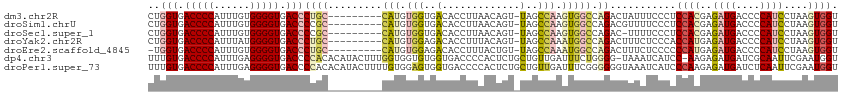
>dm3.chr2R 11787658 101 - 21146708 UCAUCUCGUGGAGGGAAAUAGUCUGGCCACUUGGCUA-ACUGUUAAGGUGUCACCACAUG---------GCAGGGUCACCCCACAAAUGGGGUCACCAG-UCAGCACCAACA--- ...((((.....))))((((((.(((((....)))))-))))))..(((((.......((---------((..(((.((((((....)))))).))).)-))))))))....--- ( -37.81, z-score = -2.02, R) >droSim1.chrU 12444360 101 - 15797150 UCAUCUCGUGGAGGGAAAACGUCUGGCCACUUGGCUA-ACUGUUAAGGUGUCACCACAUG---------GCGGGGUCACCCCACAAAUGGGGUCACCAG-CCAGCACCAACA--- ...((((.....)))).((((..(((((....)))))-..))))..(((((.......((---------((..(((.((((((....)))))).))).)-))))))))....--- ( -39.21, z-score = -1.85, R) >droSec1.super_1 9283535 100 - 14215200 UCAUCUCGUGGAGGGAAAA-GUCUGGCCACUUGGCUA-ACUGUUAAGGUGUCACCACAUG---------GCGGGGUCACCCCACAAAUGGGGUCACCAG-CCAGCACCAACA--- ...((((.....))))..(-((.(((((....)))))-))).....(((((.......((---------((..(((.((((((....)))))).))).)-))))))))....--- ( -38.41, z-score = -1.64, R) >droYak2.chr2R 3944288 97 + 21139217 UCAUCUCAUGGUGGGAGAAAGUCUGGCCAUUUGGCUA-ACUGUAAAGGUGUCUCCACAUG---------GCAGGGUCACCCCAUAAAUGGGGUCACCAG--CACCAACA------ ........((((((((((.(((.(((((....)))))-))).........))))).....---------....(((.((((((....)))))).)))..--)))))...------ ( -36.20, z-score = -1.86, R) >droEre2.scaffold_4845 8518659 91 + 22589142 UCAUCUCAUGGGGGGAGAAAGUCUGGCCAUUUGGCUA-ACAGUAAAGGUGUCUCCACAUG---------GCAGGGUCACCCCACAAAUGGGGUCACCAA--CA------------ .....(((((.(((((.(....((((((....)))..-.)))......).))))).))))---------)...(((.((((((....)))))).)))..--..------------ ( -31.90, z-score = -1.26, R) >dp4.chr3 17847152 113 - 19779522 UCAUCUCUU-GGAUGAUUUA-CCCCAGAAAUCAACAGCAGAGUGGGGUCACCACACCACCAAAGUAUGUGUGGGGUCACCCCUCAAAUGGGGUCACAAAUGUAGGAAUUCCAGCG (((((....-.)))))...(-(((((....((.......)).))))))..((((((.((....))..)))))).((.(((((......))))).))....((.((....)).)). ( -34.80, z-score = -0.80, R) >droPer1.super_73 63954 115 + 267513 UCAUCUCUUGGGAUGAUUUACCCCCCGAAAUCAACAGCAGAGUGGGGUCACCACUCCACAAAAGUAUGUGUGGGGUCACCCCUCAAAUGGGGUCACAAAUGUAGGAACUCCAGCG ((((((....))))))......(((((............((((((.....))))))((((......)))))))))..(((((......))))).......((.((....)).)). ( -38.00, z-score = -1.12, R) >consensus UCAUCUCGUGGAGGGAAAAAGUCUGGCCACUUGGCUA_ACUGUUAAGGUGUCACCACAUG_________GCGGGGUCACCCCACAAAUGGGGUCACCAG__CAGCAACAACA___ ...((((.....))))....((.(((((....))))).)).................................(((.(((((......))))).))).................. (-15.92 = -17.74 + 1.82)
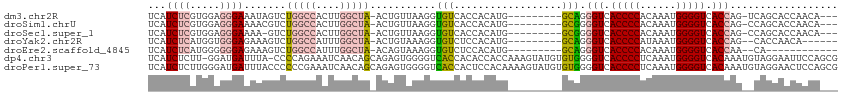
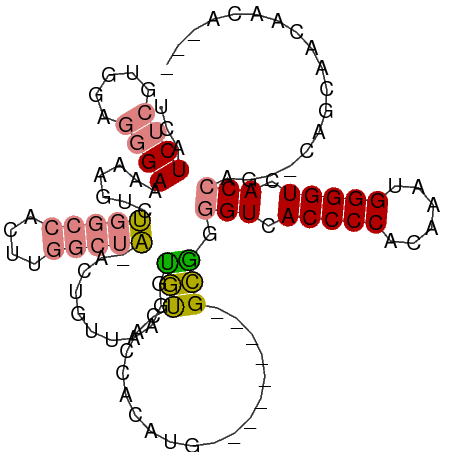
| Location | 11,787,670 – 11,787,775 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.48 |
| Shannon entropy | 0.49740 |
| G+C content | 0.52834 |
| Mean single sequence MFE | -38.46 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11787670 105 + 21146708 CUGGUGACCCCAUUUGUGGGGUGACCCUGC---------CAUGUGGUGACACCUUAACAGU-UAGCCAAGUGGCCAGACUAUUUCCCUCCACGAGAUGACCCCAUCCUAAGUGGU ..(((.((((((....)))))).)))((((---------(((.(((((((.........))-)).))).)))).)))...........((((..((((....))))....)))). ( -37.50, z-score = -2.16, R) >droSim1.chrU 12444372 105 + 15797150 CUGGUGACCCCAUUUGUGGGGUGACCCCGC---------CAUGUGGUGACACCUUAACAGU-UAGCCAAGUGGCCAGACGUUUUCCCUCCACGAGAUGACCCCAUCCUAAGUGGU ..(((.((((((....)))))).)))..((---------(((.(((((((.........))-)).))).)))))..............((((..((((....))))....)))). ( -36.80, z-score = -1.87, R) >droSec1.super_1 9283547 104 + 14215200 CUGGUGACCCCAUUUGUGGGGUGACCCCGC---------CAUGUGGUGACACCUUAACAGU-UAGCCAAGUGGCCAGAC-UUUUCCCUCCACGAGAUGACCCCAUCCUAAGUGGU ..(((.((((((....)))))).)))..((---------(((.(((((((.........))-)).))).))))).....-........((((..((((....))))....)))). ( -36.80, z-score = -2.05, R) >droYak2.chr2R 3944296 105 - 21139217 CUGGUGACCCCAUUUAUGGGGUGACCCUGC---------CAUGUGGAGACACCUUUACAGU-UAGCCAAAUGGCCAGACUUUCUCCCACCAUGAGAUGACCCCAUCCUAAGUGGU ..(((.((((((....)))))).)))((((---------(((.(((.(((.........))-)..))).)))).)))........((((...(.((((....)))))...)))). ( -35.40, z-score = -2.07, R) >droEre2.scaffold_4845 8518662 104 - 22589142 -UGGUGACCCCAUUUGUGGGGUGACCCUGC---------CAUGUGGAGACACCUUUACUGU-UAGCCAAAUGGCCAGACUUUCUCCCCCCAUGAGAUGACCCCAUCCUAAGUGGU -.(((.((((((....)))))).)))((((---------(((.(((.((((.......)))-)..))).)))).)))...........((((..((((....))))....)))). ( -35.40, z-score = -2.12, R) >dp4.chr3 17847168 113 + 19779522 UUUGUGACCCCAUUUGAGGGGUGACCCCACACAUACUUUGGUGGUGUGGUGACCCCACUCUGCUGUUGAUUUCUGGGG-UAAAUCAUCC-AAGAGAUGAUCGCAAUUCGAAUGGU .........(((((((((((((.(((.(((.(((......)))))).))).)))))....(((.(((.((((((.(((-(.....))))-.))))))))).)))..)))))))). ( -43.20, z-score = -2.52, R) >droPer1.super_73 63970 115 - 267513 UUUGUGACCCCAUUUGAGGGGUGACCCCACACAUACUUUUGUGGAGUGGUGACCCCACUCUGCUGUUGAUUUCGGGGGGUAAAUCAUCCCAAGAGAUGAUCUCAAUUCGAAUGGU .........(((((((((((((((((((............(..((((((.....))))))..)............))))....)))))))..(((.....)))...)))))))). ( -44.15, z-score = -2.30, R) >consensus CUGGUGACCCCAUUUGUGGGGUGACCCCGC_________CAUGUGGUGACACCUUAACAGU_UAGCCAAGUGGCCAGACUUUUUCCCCCCACGAGAUGACCCCAUCCUAAGUGGU ..(((.(((((......))))).))).............(((.(((..(.((.......)).)..))).)))................((((..((((....))))....)))). (-19.82 = -20.10 + 0.28)
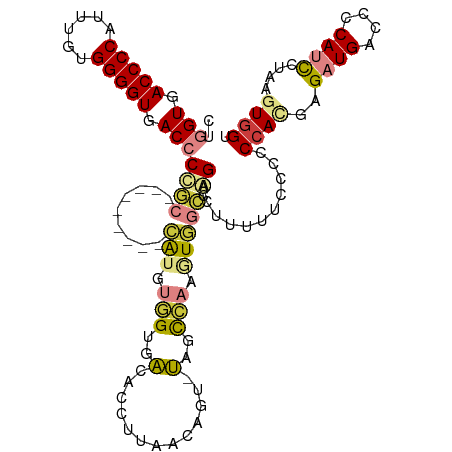
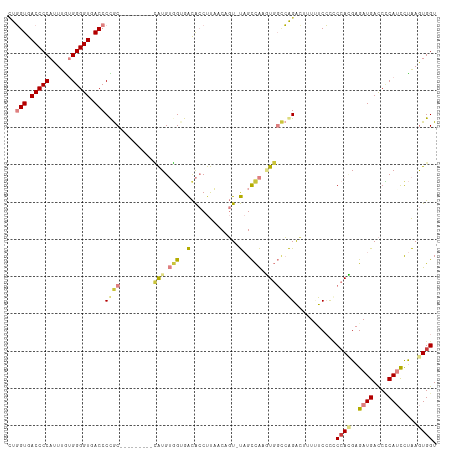
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:55 2011