| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,765,993 – 11,766,084 |
| Length | 91 |
| Max. P | 0.990247 |

| Location | 11,765,993 – 11,766,084 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 67.13 |
| Shannon entropy | 0.65058 |
| G+C content | 0.44829 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -8.69 |
| Energy contribution | -9.19 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11765993 91 - 21146708 ---------CAAAGGGAGAAUACGUG-GCAGUGGCCAUGU--GUGUGUGUGUGU--GC-GCCUUCGAUGAUUUAAUUAAUUAAAAUUGAGUUCAAUGGAGAGCGCC-- ---------..........(((((((-((....)))))))--))..........--((-(((((((.((((((((((......))))))).))).))))).)))).-- ( -29.60, z-score = -3.53, R) >droSim1.chr2R 10508309 96 - 19596830 -----CACACAAACCCAAAAGGGAGA-AUACACACUGCCA-UGUGUGUGUGUGU--GU-GCCUUCGAUGAUUUAAUUAAUUAAAAUUGAGUUCAAUGGAGAGCGCC-- -----.((((...(((....)))...-((((((((.....-.))))))))))))--((-(((((((.((((((((((......))))))).))).))))).)))).-- ( -33.70, z-score = -4.71, R) >droSec1.super_1 9260287 94 - 14215200 -----CACACAAAGCCAAAACGGAGA-AUACACACUGCCA---UGUGUGUGUGU--GC-GCCUUCGAUGAUUUAAUUAAUUAAAAUUGAGUUCAAUGGAGAGCGCC-- -----.((((....((.....))...-..((((((.....---.))))))))))--((-(((((((.((((((((((......))))))).))).))))).)))).-- ( -29.50, z-score = -3.57, R) >droEre2.scaffold_4845 8496791 90 + 22589142 -----CACACAAAGCCAAAAGGGAGA-ACACACACUGCCA-------UGUGUGU--GC-GCCUUCGAUGAUUUAAUUAAUUAAAAUUGAGUUCAAUGGAGAGCGCC-- -----.........((....))....-(((((((......-------)))))))--((-(((((((.((((((((((......))))))).))).))))).)))).-- ( -25.90, z-score = -2.88, R) >dp4.chr3 17824808 108 - 19779522 GAACCAGACCAGAGAGAGAGGGAGAGGGAGCACCCUGCCCGCCUGCCCGCCUGCUGGCUGCCUUCGAUGAUUUAAUUAAUUAAAAUUGAGUUCAAUGAAGAGACUGCU ....(((.((((..((.(.(((..((((.((.....)).).))).))).))).))))))).(((((.((((((((((......))))))).))).)))))........ ( -30.80, z-score = -1.57, R) >droPer1.super_73 41607 108 + 267513 GAACCAGACCAGAGAGAGAGGGAGAGAGAGCACCCUGCCCGCCUGCCCGCCUGCUGGCUGCCUUCGAUGAUUUAAUUAAUUAAAAUUGAGUUCAAUGAAGAGACUGCU ....(((.((((..((.(.(((((.(.(.((.....)).).))).))).))).))))))).(((((.((((((((((......))))))).))).)))))........ ( -27.80, z-score = -1.65, R) >droVir3.scaffold_12875 4444457 82 - 20611582 ---------------------GCACACACACACACACACA-UGUGUGAGUGUGC--GCUGCCUUCGAUGAUUUAAUUAAUUAAAAUUGAGUUCAAUAAAGAGCGCA-- ---------------------((((((.((((((......-)))))).))))))--((.(((((.(.((((((((((......))))))).))).).))).)))).-- ( -28.20, z-score = -4.04, R) >droGri2.scaffold_15245 17822536 100 + 18325388 --AACUACCCCCCGAGAAAACACACACGGACACACACACA-AUUGCGAGUGUGC--GCUGCCUUCGAUGAUUUAAUUAAUUAAAAUCGAGUUCAAUAAAAAGCGC--- --.........(((.(........).)))((((.(.((..-..)).).))))((--(((...((((((..(((((....)))))))))))..........)))))--- ( -15.02, z-score = -0.24, R) >consensus _____CACACAAAGAGAAAAGGCAGA_ACACACACUGCCA__GUGUGUGUGUGC__GC_GCCUUCGAUGAUUUAAUUAAUUAAAAUUGAGUUCAAUGAAGAGCGCC__ ...........................................................(((((((.((((((((((......))))))).))).))))).))..... ( -8.69 = -9.19 + 0.50)


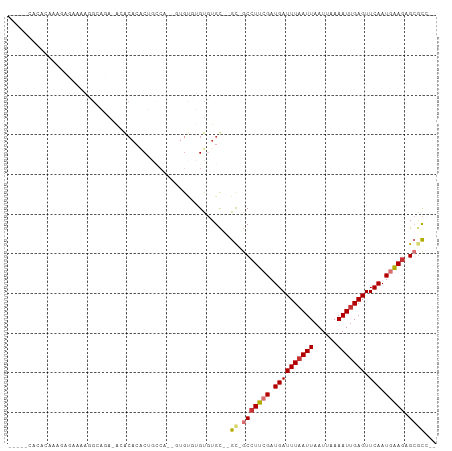
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:51 2011