
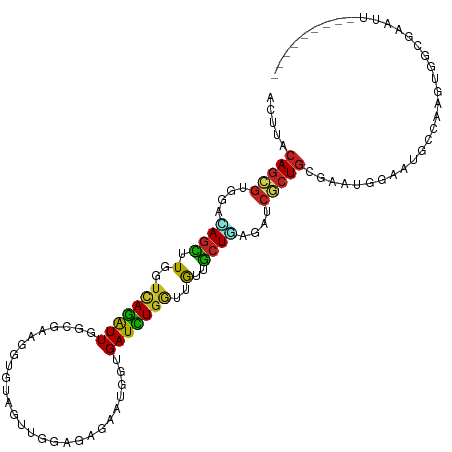
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,764,529 – 11,764,635 |
| Length | 106 |
| Max. P | 0.594139 |

| Location | 11,764,529 – 11,764,635 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.20 |
| Shannon entropy | 0.65858 |
| G+C content | 0.45043 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -13.77 |
| Energy contribution | -13.26 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.57 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11764529 106 + 21146708 ACUUACAGCGUGGACAGCUUGGUCAGAUUGGCAAAGGUGUAGUUGGAGAGAAUGGUGAUCUGGUUGUUGCUGAGAUCGCUGCGAAUGGAAUGCCAAGUGGCGAAUU--------- .(((.((((....(((.(((.(((.....))).))).))).))))..)))...(((((((((((....))).))))))))..........((((....))))....--------- ( -30.30, z-score = -0.77, R) >apiMel3.Group8 7181717 111 - 11452794 ACUUACAAGUGGGACAAUUUGGUGAGAUUUCUGAAGGUAUCGUUGGACAGCAUCCCGAUCUGAUUGUUGCUCAGAUCCCUGGAAAAAAAAAAAGAAAGGGAGGGAAAUCUU---- .....(((((......)))))..(((((((((.........(((....))).(((((((((((.......))))))).((............))...)))).)))))))))---- ( -25.50, z-score = 0.38, R) >anoGam1.chr2R 35100321 111 - 62725911 ACUUACAGUGUGGAGAGUUUGCUUAGAUUUGCAAAGGUGUGGUUGGACAGUAUUCCGAUCUGGUUGUUGCUCAAAUCCCUGUGGGGGGAAAAUGGUAAAAGAUUGCACAUU---- ......((((((.((..((((((.......((((......(((((((......)))))))......)))).....(((((....)))))....))))))...)).))))))---- ( -28.30, z-score = -0.24, R) >droGri2.scaffold_15245 17820321 115 - 18325388 ACUUACAGCGUGGAUAGUUUGGUCAGAUUGGCAAAGGUGUAAUUGGAUAGUAUGGUGAUUUGGUUGUUGCUAAGAUCGCUGCAAAGAAAAAAGAACAGCACAAAAAAAAAAGUGG .(((.(((((....((((.(..((((((((.((..(.(((......))).).)).))))))))..)..))))....)))))..)))............(((..........))). ( -19.40, z-score = 0.87, R) >droVir3.scaffold_12875 4442301 101 + 20611582 ACUUACAGCGUGGAUAGCUUGGUCAGAUUGGCAAAGGUGUAAUUGGACAGUAUGGUGAUUUGAUUGUUGCUGAGAUCGCUGCAAAUGAAGCAAAACAAUUU-------------- .....(((((....((((.(((((((((((.((..(.(((......))).).)).)))))))))))..))))....)))))....................-------------- ( -22.30, z-score = 0.26, R) >dp4.chr3 17823359 82 + 19779522 ACUUACAGCGUGGAUAGUUUGGUCAGAUUGGCGAAGGUGUUGUUGGAGAGUAUGGUGAUCUGGUUGUUGCUCAGAUCGCUGU--------------------------------- ((((.((((...((((.(((.(((.....))).))).))))))))..))))(((((((((((((....)).)))))))))))--------------------------------- ( -26.40, z-score = -2.51, R) >droEre2.scaffold_4845 8495374 106 - 22589142 ACUUACAGCGUGGACAGCUUGGUCAGAUUGGCGAAGGUGUAGUUGGAGAGAAUGGUGAUCUGGUUGUUGCUGAGAUCGCUGCGAAUGUAAUGCCAAGUGGCGAAUU--------- .(((.((((....(((.(((.(((.....))).))).))).))))..)))...(((((((((((....))).))))))))..........((((....))))....--------- ( -32.40, z-score = -1.40, R) >droYak2.chr2R 3920452 106 - 21139217 ACUUACAGCGUGGACAGCUUGGUCAGAUUGGCAAAGGUGUAGUUGGAGAGAAUGGUGAUCUGGUUGUUGCUGAGAUCCCUGCGAAUGGAAUGCCAAGUGGCGAGUU--------- .(((.((((....(((.(((.(((.....))).))).))).))))..)))...((.((((((((....))).)))))))...........((((....))))....--------- ( -27.90, z-score = 0.41, R) >droSec1.super_1 9258960 106 + 14215200 ACUUACAGCGUGGACAGCUUGGUCAGAUUGGCGAAGGUGUAGUUGGAGAGAAUGGUGAUCUGGUUGUUGCUGAGAUCGCUGCGAAUGGAAUGCCAAGUGGCGAAUU--------- .(((.((((....(((.(((.(((.....))).))).))).))))..)))...(((((((((((....))).))))))))..........((((....))))....--------- ( -32.40, z-score = -1.40, R) >droSim1.chr2R 10506981 106 + 19596830 ACUUACAGCGUGGACAGCUUGGUCAGAUUGGCGAAGGUGUAGUUGGAGAGAAUGGUGAUCUGGUUGUUGCUGAGAUCGCUGCGAAUGGAAUGCCAAGUGGCGAAUU--------- .(((.((((....(((.(((.(((.....))).))).))).))))..)))...(((((((((((....))).))))))))..........((((....))))....--------- ( -32.40, z-score = -1.40, R) >triCas2.ChLG2 11919629 91 + 12900155 ------AGGGUCGAAAGUUUCGAUAAGUUGGCGAAAGUGUUGUUUGAUAACAUCCCGAUUUGGUUACUACUCAAAUCCCUAAAAUAAUCAUCCAAUU------------------ ------((((.....(((.....((((((((.((...((((((...))))))))))))))))......))).....)))).................------------------ ( -17.00, z-score = -0.46, R) >consensus ACUUACAGCGUGGACAGCUUGGUCAGAUUGGCGAAGGUGUAGUUGGAGAGAAUGGUGAUCUGGUUGUUGCUGAGAUCGCUGCGAAUGGAAUGCCAAGUGGCGAAUU_________ .....(((((....((((.(..(((((((...........................)))))))..)..))))....))))).................................. (-13.77 = -13.26 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:50 2011