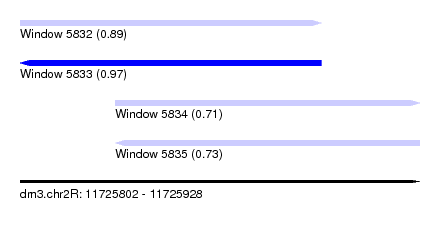
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,725,802 – 11,725,928 |
| Length | 126 |
| Max. P | 0.965318 |

| Location | 11,725,802 – 11,725,897 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 84.26 |
| Shannon entropy | 0.29204 |
| G+C content | 0.45441 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -16.14 |
| Energy contribution | -16.61 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
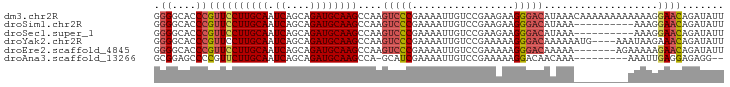
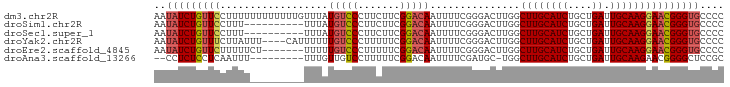
>dm3.chr2R 11725802 95 + 21146708 GGGGCACCCGUUCCUUGCAAUCAGCAGAUGCAAGCCAAGUCCCGAAAAUUGUCCGAAGAAGGGACAUAAACAAAAAAAAAAAAGGAACAGAUAUU (((...)))((((((((((.((....))))).......(((((.................)))))................)))))))....... ( -23.33, z-score = -2.23, R) >droSim1.chr2R 10465482 85 + 19596830 GGGGCACCCGUUCCUUGCAAUCAGCAGAUGCAAGCCAAGUCCCGAAAAUUGUCCGAAGAAGGGACAUAAA----------AAAGGAACAGAUAUU (((...)))((((((((((.((....))))).......(((((.................))))).....----------.)))))))....... ( -23.33, z-score = -2.06, R) >droSec1.super_1 9219952 85 + 14215200 GGGGCACCCGUUCCUUGCAAUCAGCAGAUGCAAGCCAAGUCCCGAAAAUUGUCCGAAGAAGGGACAUAAA----------AAAGGAACAGAUAUU (((...)))((((((((((.((....))))).......(((((.................))))).....----------.)))))))....... ( -23.33, z-score = -2.06, R) >droYak2.chr2R 3879806 91 - 21139217 GGGGCACCCGUUCCUUGCAAUCAGCAGAUGCAAGCCAAGUCCCGAAAAUUGUCCGAAAAAGGGACAAAAAAUG----AAAUAAGAAACAGAUAUU (((((........((((((.((....))))))))....))))).....((((((.......))))))......----.................. ( -21.00, z-score = -1.66, R) >droEre2.scaffold_4845 8455330 88 - 22589142 GGGGCACCCGUUCCUUGCAAUCAGCAGAUGCAAGCCAAGUCCCGAAAAUUGUCCGAAAAAGGGACAAAAA-------AGAAAAAGAACAGAUAUU (((...)))((((((((((.((....))))))))....(((((.................))))).....-------.......))))....... ( -21.63, z-score = -2.09, R) >droAna3.scaffold_13266 7414169 83 - 19884421 GCGGAGCCCCGUUCUUGCAAUCAGCAGAUGCAAGCCA-GCAUCGAAAAUUGUCCGAAAAAGGACAACAAA---------AAAUUGAGGAGAGG-- ((((....))))((((.((((.....(((((......-))))).....((((((......))))))....---------..))))..))))..-- ( -22.80, z-score = -2.03, R) >consensus GGGGCACCCGUUCCUUGCAAUCAGCAGAUGCAAGCCAAGUCCCGAAAAUUGUCCGAAAAAGGGACAUAAA_________AAAAGGAACAGAUAUU .((....))((((((((((.((....))))))))....(((((.................)))))...................))))....... (-16.14 = -16.61 + 0.47)
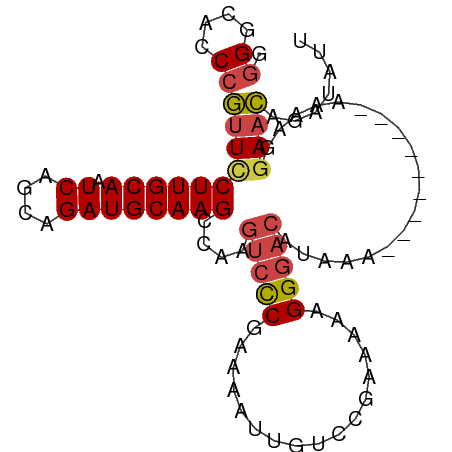
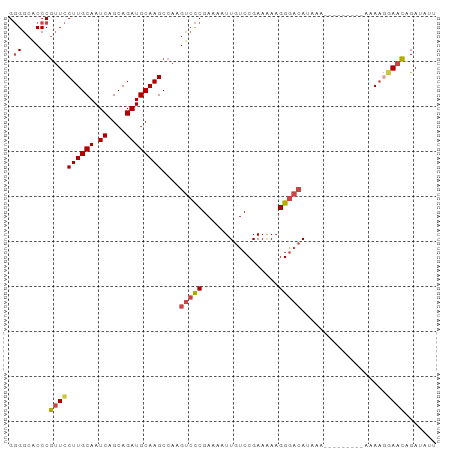
| Location | 11,725,802 – 11,725,897 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 84.26 |
| Shannon entropy | 0.29204 |
| G+C content | 0.45441 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11725802 95 - 21146708 AAUAUCUGUUCCUUUUUUUUUUUUGUUUAUGUCCCUUCUUCGGACAAUUUUCGGGACUUGGCUUGCAUCUGCUGAUUGCAAGGAACGGGUGCCCC ..((((((((((............(((...(((((.((....))........)))))..)))(((((((....)).))))))))))))))).... ( -27.60, z-score = -2.82, R) >droSim1.chr2R 10465482 85 - 19596830 AAUAUCUGUUCCUUU----------UUUAUGUCCCUUCUUCGGACAAUUUUCGGGACUUGGCUUGCAUCUGCUGAUUGCAAGGAACGGGUGCCCC ..((((((((((((.----------.....(((((.((....))........)))))(..((........))..)....)))))))))))).... ( -26.20, z-score = -2.50, R) >droSec1.super_1 9219952 85 - 14215200 AAUAUCUGUUCCUUU----------UUUAUGUCCCUUCUUCGGACAAUUUUCGGGACUUGGCUUGCAUCUGCUGAUUGCAAGGAACGGGUGCCCC ..((((((((((((.----------.....(((((.((....))........)))))(..((........))..)....)))))))))))).... ( -26.20, z-score = -2.50, R) >droYak2.chr2R 3879806 91 + 21139217 AAUAUCUGUUUCUUAUUU----CAUUUUUUGUCCCUUUUUCGGACAAUUUUCGGGACUUGGCUUGCAUCUGCUGAUUGCAAGGAACGGGUGCCCC ..................----......((((((.......)))))).....((((((((.((((((((....)).))))))...))))).))). ( -23.80, z-score = -1.72, R) >droEre2.scaffold_4845 8455330 88 + 22589142 AAUAUCUGUUCUUUUUCU-------UUUUUGUCCCUUUUUCGGACAAUUUUCGGGACUUGGCUUGCAUCUGCUGAUUGCAAGGAACGGGUGCCCC ..(((((((((...((((-------...((((((.......)))))).....)))).....((((((((....)).))))))))))))))).... ( -25.00, z-score = -2.31, R) >droAna3.scaffold_13266 7414169 83 + 19884421 --CCUCUCCUCAAUUU---------UUUGUUGUCCUUUUUCGGACAAUUUUCGAUGC-UGGCUUGCAUCUGCUGAUUGCAAGAACGGGGCUCCGC --.....((((.....---------...(((((((......))))))).((((((((-......)))))(((.....))).))).))))...... ( -23.10, z-score = -1.86, R) >consensus AAUAUCUGUUCCUUUU_________UUUAUGUCCCUUCUUCGGACAAUUUUCGGGACUUGGCUUGCAUCUGCUGAUUGCAAGGAACGGGUGCCCC ..(((((((((..................(((((.......)))))...............((((((((....)).))))))))))))))).... (-19.33 = -19.80 + 0.47)
| Location | 11,725,832 – 11,725,928 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.63 |
| Shannon entropy | 0.39810 |
| G+C content | 0.37032 |
| Mean single sequence MFE | -15.08 |
| Consensus MFE | -7.60 |
| Energy contribution | -8.96 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
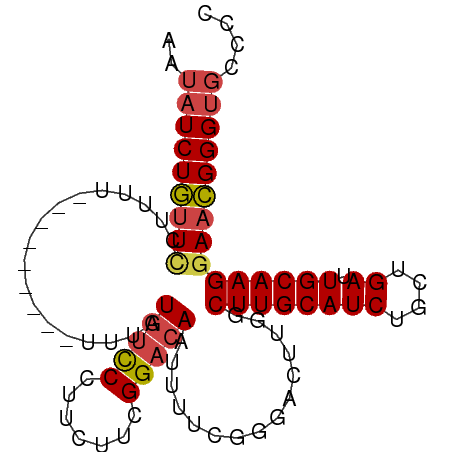
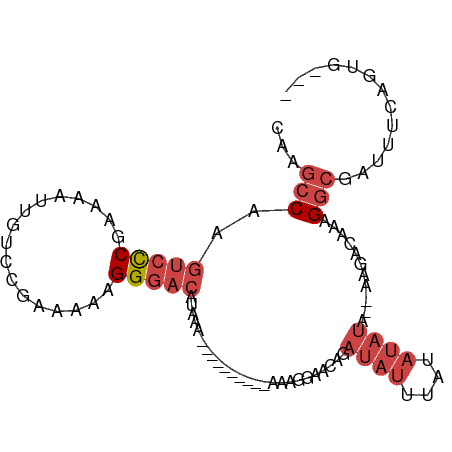
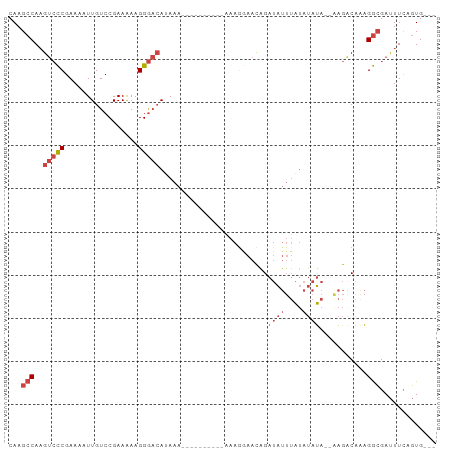
>dm3.chr2R 11725832 96 + 21146708 CAAGCCAAGUCCCGAAAAUUGUCCGAAGAAGGGACAUAAACAAAAAAAAAAAAGGAACAGAUAUUUAUAUAUAUAAAGACAAAGGCGAUUUCAGUG--- ...(((..(((((.................)))))............................(((((....)))))......)))..........--- ( -13.83, z-score = -1.84, R) >droSim1.chr2R 10465512 84 + 19596830 CAAGCCAAGUCCCGAAAAUUGUCCGAAGAAGGGACAUAAA----------AAAGGAACAGAUAUUUAUAUAUA--AAGACAAAGGCGAUUUCAGUG--- ...(((..((.((......(((((.......)))))....----------...)).))..((((....)))).--........)))..........--- ( -15.02, z-score = -2.20, R) >droSec1.super_1 9219982 84 + 14215200 CAAGCCAAGUCCCGAAAAUUGUCCGAAGAAGGGACAUAAA----------AAAGGAACAGAUAUUUAUAUAUA--AAGACAAAGGCGAUUUCAGUG--- ...(((..((.((......(((((.......)))))....----------...)).))..((((....)))).--........)))..........--- ( -15.02, z-score = -2.20, R) >droYak2.chr2R 3879836 90 - 21139217 CAAGCCAAGUCCCGAAAAUUGUCCGAAAAAGGGACAAAAA----AUGAAAUAAGAAACAGAUAUUUAUAUAUA--GAGGCAGAGGCGAUUUCAGUG--- ...(((..(((.(.....((((((.......))))))...----................((((....)))).--).)))...)))..........--- ( -13.80, z-score = -1.15, R) >droEre2.scaffold_4845 8455360 87 - 22589142 CAAGCCAAGUCCCGAAAAUUGUCCGAAAAAGGGACAAAAA-------AGAAAAAGAACAGAUAUUUAUAUAUA--AAGACAAAGGCGAUUUCAGUG--- ...(((..(((.......((((((.......))))))...-------.............((((....)))).--..)))...)))..........--- ( -13.80, z-score = -2.31, R) >droAna3.scaffold_13266 7414199 82 - 19884421 CAAGCCA-GCAUCGAAAAUUGUCCGAAAAAGGACAACAAA--------------AAAUUGAGGAGAGGAGACA--AGGACCAGGAGGACUUUGUUGAUG .......-.((((.....((((((......))))))....--------------...............((((--(((.((....)).))))))))))) ( -19.00, z-score = -2.37, R) >consensus CAAGCCAAGUCCCGAAAAUUGUCCGAAAAAGGGACAUAAA__________AAAGGAACAGAUAUUUAUAUAUA__AAGACAAAGGCGAUUUCAGUG___ ...(((..(((((.................))))).........................((((....))))...........)))............. ( -7.60 = -8.96 + 1.36)
| Location | 11,725,832 – 11,725,928 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.63 |
| Shannon entropy | 0.39810 |
| G+C content | 0.37032 |
| Mean single sequence MFE | -14.49 |
| Consensus MFE | -7.81 |
| Energy contribution | -8.65 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.729958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
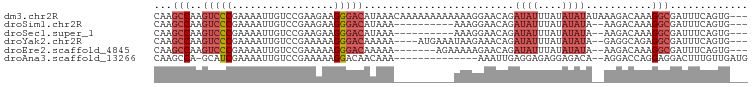
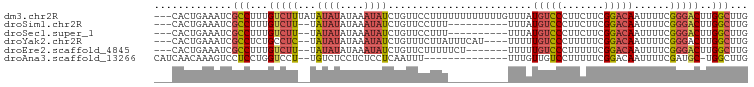
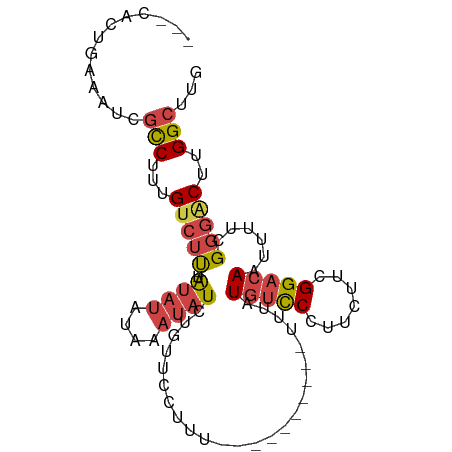
>dm3.chr2R 11725832 96 - 21146708 ---CACUGAAAUCGCCUUUGUCUUUAUAUAUAUAAAUAUCUGUUCCUUUUUUUUUUUUGUUUAUGUCCCUUCUUCGGACAAUUUUCGGGACUUGGCUUG ---..........(((...(((((.......(((((((...................)))))))((((.......)))).......)))))..)))... ( -15.31, z-score = -2.02, R) >droSim1.chr2R 10465512 84 - 19596830 ---CACUGAAAUCGCCUUUGUCUU--UAUAUAUAAAUAUCUGUUCCUUU----------UUUAUGUCCCUUCUUCGGACAAUUUUCGGGACUUGGCUUG ---..........((((((((...--.....))))).....(((((...----------....(((((.......)))))......)))))..)))... ( -14.12, z-score = -1.74, R) >droSec1.super_1 9219982 84 - 14215200 ---CACUGAAAUCGCCUUUGUCUU--UAUAUAUAAAUAUCUGUUCCUUU----------UUUAUGUCCCUUCUUCGGACAAUUUUCGGGACUUGGCUUG ---..........((((((((...--.....))))).....(((((...----------....(((((.......)))))......)))))..)))... ( -14.12, z-score = -1.74, R) >droYak2.chr2R 3879836 90 + 21139217 ---CACUGAAAUCGCCUCUGCCUC--UAUAUAUAAAUAUCUGUUUCUUAUUUCAU----UUUUUGUCCCUUUUUCGGACAAUUUUCGGGACUUGGCUUG ---..........(((....(((.--.......(((((.........)))))...----...((((((.......)))))).....)))....)))... ( -13.30, z-score = -1.21, R) >droEre2.scaffold_4845 8455360 87 + 22589142 ---CACUGAAAUCGCCUUUGUCUU--UAUAUAUAAAUAUCUGUUCUUUUUCU-------UUUUUGUCCCUUUUUCGGACAAUUUUCGGGACUUGGCUUG ---..........(((...(((((--.((((....)))).............-------...((((((.......)))))).....)))))..)))... ( -14.20, z-score = -2.05, R) >droAna3.scaffold_13266 7414199 82 + 19884421 CAUCAACAAAGUCCUCCUGGUCCU--UGUCUCCUCUCCUCAAUUU--------------UUUGUUGUCCUUUUUCGGACAAUUUUCGAUGC-UGGCUUG ((((.((((.(.((....)).).)--)))................--------------...(((((((......)))))))....)))).-....... ( -15.90, z-score = -2.44, R) >consensus ___CACUGAAAUCGCCUUUGUCUU__UAUAUAUAAAUAUCUGUUCCUUU__________UUUAUGUCCCUUCUUCGGACAAUUUUCGGGACUUGGCUUG .............(((...(((((...((((....))))........................(((((.......)))))......)))))..)))... ( -7.81 = -8.65 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:42 2011