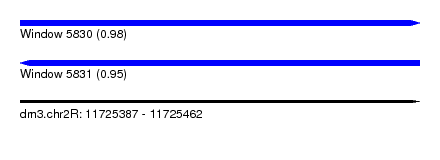
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,725,387 – 11,725,462 |
| Length | 75 |
| Max. P | 0.984666 |

| Location | 11,725,387 – 11,725,462 |
|---|---|
| Length | 75 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 68.21 |
| Shannon entropy | 0.53394 |
| G+C content | 0.50213 |
| Mean single sequence MFE | -23.39 |
| Consensus MFE | -13.95 |
| Energy contribution | -15.46 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
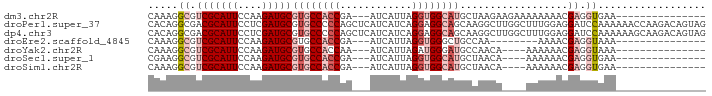
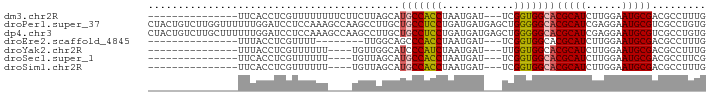
>dm3.chr2R 11725387 75 + 21146708 CAAAGGCGUCGCAUUCCAAGAUGCGUGCCACCGA---AUCAUUAGGUGGCAUGCUAAGAAGAAAAAAAACGAGGUGAA--------------- .....((.(((..(((......((((((((((..---.......))))))))))......)))......))).))...--------------- ( -23.60, z-score = -2.99, R) >droPer1.super_37 297649 93 - 760358 CACAGGCGACGCAUUCCUCGAUGCGUGCCCCCAGCUCAUCAUCAGGAGGCAGCAAGGCUUGGCUUUGGAGGAUCCAAAAAACCAAGACAGUAG ....(((.(((((((....))))))))))((..((((.((.....)).).)))..))(((((.(((((.....)))))...)))))....... ( -27.00, z-score = -0.42, R) >dp4.chr3 318675 93 - 19779522 CACAGGCGACGCAUUCCUCGAUGCGUGCCCCCAGCUCAUCAUCAGGAGGCAGCAAGGCUUGGCUUUGGAGGAUCCAAAAAAGCAAGACAGUAG ..(((((.(((((((....)))))))(((.((............)).)))......)))))(((((((.....)))...)))).......... ( -26.70, z-score = -0.11, R) >droEre2.scaffold_4845 8454934 67 - 22589142 CAAAGGCGUCGCAUUCCAAGAUGCGUGCCACCGA---AUCAUUAGGUGGGCUGCCAA--------AAAACGAGGUAAA--------------- ....(((..((((((....))))))..(((((..---.......)))))...)))..--------.............--------------- ( -19.50, z-score = -1.27, R) >droYak2.chr2R 3879393 71 - 21139217 CAAAGGCGUCGCAUUCCAAGAUGCGUGCCACCAA---AUCAUUAGAUGGGAUGCCAACA----AAAAAACGAGGUAAA--------------- ....((((.((((((....))))))))))(((..---(((....)))((....))....----.........)))...--------------- ( -17.60, z-score = -1.74, R) >droSec1.super_1 9219542 71 + 14215200 CGAAGGCGUCGCAUUCCAAGAUGCGUGCCACCGA---AUCAUUAGGUGGCAUGCUAACA----AAAAAACGAGGUGAA--------------- .....((.(((...........((((((((((..---.......)))))))))).....----......))).))...--------------- ( -24.65, z-score = -2.98, R) >droSim1.chr2R 10465073 71 + 19596830 CAAAGGCGUCGCAUUCCAAGAUGCGUGCCACCGA---AUCAUUAGGUGGCAUGCUAACA----AAAAAACGAGGUGAA--------------- .....((.(((...........((((((((((..---.......)))))))))).....----......))).))...--------------- ( -24.65, z-score = -3.32, R) >consensus CAAAGGCGUCGCAUUCCAAGAUGCGUGCCACCGA___AUCAUUAGGUGGCAUGCUAACA____AAAAAACGAGGUAAA_______________ .....((.(((((((....)))))((((((((............))))))))..................)).)).................. (-13.95 = -15.46 + 1.51)
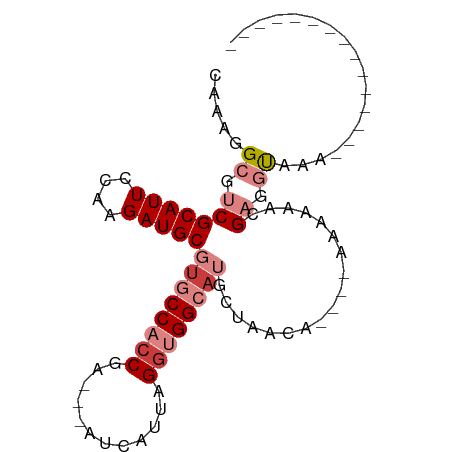
| Location | 11,725,387 – 11,725,462 |
|---|---|
| Length | 75 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 68.21 |
| Shannon entropy | 0.53394 |
| G+C content | 0.50213 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -14.70 |
| Energy contribution | -14.60 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
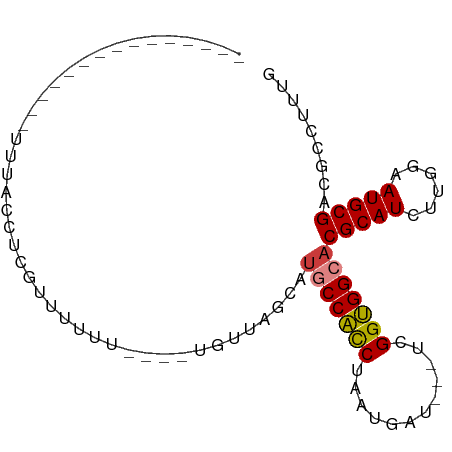
>dm3.chr2R 11725387 75 - 21146708 ---------------UUCACCUCGUUUUUUUUCUUCUUAGCAUGCCACCUAAUGAU---UCGGUGGCACGCAUCUUGGAAUGCGACGCCUUUG ---------------......((((.....((((.....((.(((((((.......---..))))))).)).....)))).))))........ ( -20.00, z-score = -2.72, R) >droPer1.super_37 297649 93 + 760358 CUACUGUCUUGGUUUUUUGGAUCCUCCAAAGCCAAGCCUUGCUGCCUCCUGAUGAUGAGCUGGGGGCACGCAUCGAGGAAUGCGUCGCCUGUG .......((((((..(((((.....)))))))))))(((.(((.(.((.....)).)))).)))(((((((((......)))))).))).... ( -29.90, z-score = -0.50, R) >dp4.chr3 318675 93 + 19779522 CUACUGUCUUGCUUUUUUGGAUCCUCCAAAGCCAAGCCUUGCUGCCUCCUGAUGAUGAGCUGGGGGCACGCAUCGAGGAAUGCGUCGCCUGUG .(((.((..(((..((((((.....)))))).....(((((((((((((............))))))).)))...)))...)))..))..))) ( -25.50, z-score = 0.70, R) >droEre2.scaffold_4845 8454934 67 + 22589142 ---------------UUUACCUCGUUUU--------UUGGCAGCCCACCUAAUGAU---UCGGUGGCACGCAUCUUGGAAUGCGACGCCUUUG ---------------.............--------..(((.(((.(((.......---..)))))).(((((......)))))..))).... ( -18.10, z-score = -1.40, R) >droYak2.chr2R 3879393 71 + 21139217 ---------------UUUACCUCGUUUUUU----UGUUGGCAUCCCAUCUAAUGAU---UUGGUGGCACGCAUCUUGGAAUGCGACGCCUUUG ---------------...............----....(((...(((((.......---..)))))..(((((......)))))..))).... ( -15.30, z-score = -0.61, R) >droSec1.super_1 9219542 71 - 14215200 ---------------UUCACCUCGUUUUUU----UGUUAGCAUGCCACCUAAUGAU---UCGGUGGCACGCAUCUUGGAAUGCGACGCCUUCG ---------------......((((.((((----.....((.(((((((.......---..))))))).)).....)))).))))........ ( -19.60, z-score = -1.86, R) >droSim1.chr2R 10465073 71 - 19596830 ---------------UUCACCUCGUUUUUU----UGUUAGCAUGCCACCUAAUGAU---UCGGUGGCACGCAUCUUGGAAUGCGACGCCUUUG ---------------......((((.((((----.....((.(((((((.......---..))))))).)).....)))).))))........ ( -19.60, z-score = -1.98, R) >consensus _______________UUUACCUCGUUUUUU____UGUUAGCAUGCCACCUAAUGAU___UCGGUGGCACGCAUCUUGGAAUGCGACGCCUUUG ..........................................(((((((............)))))))(((((......)))))......... (-14.70 = -14.60 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:40 2011