
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,685,335 – 11,685,426 |
| Length | 91 |
| Max. P | 0.972016 |

| Location | 11,685,335 – 11,685,426 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.68 |
| Shannon entropy | 0.34321 |
| G+C content | 0.48511 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -22.95 |
| Energy contribution | -23.19 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
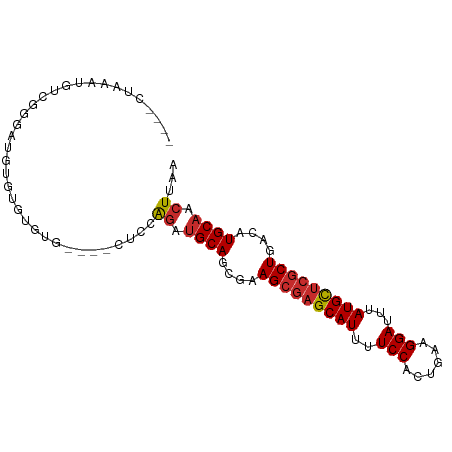
>dm3.chr2R 11685335 91 + 21146708 ----CUAAAUGUCGGGAUGUGUGUGUGUGUGCUCCAAAUGCAGCGAAGCGAGCAUUUUCCACUGAAGGAUUUAUGCUCGCUGACAUGCAACUUAA ----.........(((.(((((((.(((.(((.......)))))).(((((((((..(((......)))...))))))))).))))))).))).. ( -29.80, z-score = -2.11, R) >dp4.chr3 279672 92 - 19779522 GUGUAUGCAGCUCCGGCUCUGGCUCUGG---CUCUGGGUGCAGCGGAGCGAGCAUUUUCCACUGAAGGAUUUAUGCUCGCUGACAUGCAACUUAA .((((((((((....((((((((....)---).(((....)))))))))((((((..(((......)))...)))))))))).))))))...... ( -37.30, z-score = -2.23, R) >droPer1.super_37 258687 92 - 760358 GUGUAUGCAGCUCCGGCUCUGGCUCUGG---CUCUGGGUGCAGCGGAGCGAGCAUUUUCCACUGAAGGAUUUAUGCUCGCUGACAUGCAACUUAA .((((((((((....((((((((....)---).(((....)))))))))((((((..(((......)))...)))))))))).))))))...... ( -37.30, z-score = -2.23, R) >droAna3.scaffold_13266 7367319 91 - 19884421 CAGUACGUAGCUCCAGAAAUGUGUGUG----CUCCAGAUGCAGCGGAGCGAGCAUUUUCCACUGAAGGAUUUAUGCUCGCUGACAUGCAACUUAA ((((.....(((((.(...(((((.((----...)).))))).))))))((((((..(((......)))...))))))))))............. ( -29.30, z-score = -1.59, R) >droEre2.scaffold_4845 8411670 87 - 22589142 ----CUAAAUGUCGGGAUGUGUGUGUG----CUCCAGAUGCAGCGAAGCGAGCAUUUUCCACUGAAGGAUUUAUGCUCGCUGACAUGCAACUUAA ----......(((.(((.((......)----)))).)))(((....(((((((((..(((......)))...)))))))))....)))....... ( -29.80, z-score = -2.32, R) >droYak2.chr2R 3837985 89 - 21139217 ----CUAAAUGUCGGGAUGUGUGUGUGUG--CUCCAGAUGCAGCGAAGCGAGCAUUUUCCACUGAAGGAUUUAUGCUCGCUGACAUGCAACUUAA ----......(((.(((.(..(....)..--)))).)))(((....(((((((((..(((......)))...)))))))))....)))....... ( -30.60, z-score = -2.30, R) >droSec1.super_1 9179050 87 + 14215200 ----CUAAAUGUCGGGAUAUGUGUGUG----CUCCAGAUGCAGCGAAGCGAGCAUUUUCCACUGAAGGAUUUAUGCUCGCUGAAAUGCAACUUAA ----.........((..(((....)))----..))((.((((....(((((((((..(((......)))...)))))))))....)))).))... ( -29.20, z-score = -2.74, R) >droSim1.chr2R 10422549 87 + 19596830 ----CUAAAUGUCGGGAUGUGUGUGUG----CUCCAGAUGCAGCGAAGCGAGCAUUUUCCACUGAAGGAUUUAUGCUCGCUGACAUGCAACUUAA ----......(((.(((.((......)----)))).)))(((....(((((((((..(((......)))...)))))))))....)))....... ( -29.80, z-score = -2.32, R) >droVir3.scaffold_12823 1094777 87 - 2474545 ----GUGCGUGUGUGCAUGUGUGUGUA----UUUUGGGCGCACUGAAGCAAGCAUUUUCCACCGAGGGAUUUAUGUGCACUGACAUGCAACUUAA ----.(((((((((((..((((((.(.----....).))))))........((((..(((......)))...))))))))..)))))))...... ( -27.40, z-score = -0.45, R) >consensus ____CUAAAUGUCGGGAUGUGUGUGUG____CUCCAGAUGCAGCGAAGCGAGCAUUUUCCACUGAAGGAUUUAUGCUCGCUGACAUGCAACUUAA ...................................((.((((....(((((((((..(((......)))...)))))))))....)))).))... (-22.95 = -23.19 + 0.24)

| Location | 11,685,335 – 11,685,426 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 82.68 |
| Shannon entropy | 0.34321 |
| G+C content | 0.48511 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -20.35 |
| Energy contribution | -20.28 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11685335 91 - 21146708 UUAAGUUGCAUGUCAGCGAGCAUAAAUCCUUCAGUGGAAAAUGCUCGCUUCGCUGCAUUUGGAGCACACACACACACAUCCCGACAUUUAG---- .........((((((((((((((...(((......)))..)))))))))..(((.(....).))).................)))))....---- ( -27.30, z-score = -3.26, R) >dp4.chr3 279672 92 + 19779522 UUAAGUUGCAUGUCAGCGAGCAUAAAUCCUUCAGUGGAAAAUGCUCGCUCCGCUGCACCCAGAG---CCAGAGCCAGAGCCGGAGCUGCAUACAC ......((((.((.(((((((((...(((......)))..)))))))))..))))))..(((..---((.(.((....)))))..)))....... ( -28.60, z-score = -1.37, R) >droPer1.super_37 258687 92 + 760358 UUAAGUUGCAUGUCAGCGAGCAUAAAUCCUUCAGUGGAAAAUGCUCGCUCCGCUGCACCCAGAG---CCAGAGCCAGAGCCGGAGCUGCAUACAC ......((((.((.(((((((((...(((......)))..)))))))))..))))))..(((..---((.(.((....)))))..)))....... ( -28.60, z-score = -1.37, R) >droAna3.scaffold_13266 7367319 91 + 19884421 UUAAGUUGCAUGUCAGCGAGCAUAAAUCCUUCAGUGGAAAAUGCUCGCUCCGCUGCAUCUGGAG----CACACACAUUUCUGGAGCUACGUACUG ...(((((((.((.(((((((((...(((......)))..)))))))))..))))))((..(((----.........)))..)))))........ ( -26.70, z-score = -1.36, R) >droEre2.scaffold_4845 8411670 87 + 22589142 UUAAGUUGCAUGUCAGCGAGCAUAAAUCCUUCAGUGGAAAAUGCUCGCUUCGCUGCAUCUGGAG----CACACACACAUCCCGACAUUUAG---- .........((((((((((((((...(((......)))..)))))))))..(((.(....).))----).............)))))....---- ( -27.30, z-score = -3.08, R) >droYak2.chr2R 3837985 89 + 21139217 UUAAGUUGCAUGUCAGCGAGCAUAAAUCCUUCAGUGGAAAAUGCUCGCUUCGCUGCAUCUGGAG--CACACACACACAUCCCGACAUUUAG---- .........((((((((((((((...(((......)))..)))))))))..(((.(....).))--)...............)))))....---- ( -27.30, z-score = -3.09, R) >droSec1.super_1 9179050 87 - 14215200 UUAAGUUGCAUUUCAGCGAGCAUAAAUCCUUCAGUGGAAAAUGCUCGCUUCGCUGCAUCUGGAG----CACACACAUAUCCCGACAUUUAG---- ....((((......(((((((((...(((......)))..)))))))))..(((.(....).))----)............))))......---- ( -25.10, z-score = -2.91, R) >droSim1.chr2R 10422549 87 - 19596830 UUAAGUUGCAUGUCAGCGAGCAUAAAUCCUUCAGUGGAAAAUGCUCGCUUCGCUGCAUCUGGAG----CACACACACAUCCCGACAUUUAG---- .........((((((((((((((...(((......)))..)))))))))..(((.(....).))----).............)))))....---- ( -27.30, z-score = -3.08, R) >droVir3.scaffold_12823 1094777 87 + 2474545 UUAAGUUGCAUGUCAGUGCACAUAAAUCCCUCGGUGGAAAAUGCUUGCUUCAGUGCGCCCAAAA----UACACACACAUGCACACACGCAC---- ....(((((((((..(((........(((......)))...((..(((......)))..))...----..)))..))))))).))......---- ( -16.40, z-score = 0.93, R) >consensus UUAAGUUGCAUGUCAGCGAGCAUAAAUCCUUCAGUGGAAAAUGCUCGCUUCGCUGCAUCUGGAG____CACACACACAUCCCGACAUUUAG____ ......((((....(((((((((...(((......)))..)))))))))....))))...................................... (-20.35 = -20.28 + -0.07)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:34 2011