
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,966,974 – 2,967,105 |
| Length | 131 |
| Max. P | 0.984547 |

| Location | 2,966,974 – 2,967,105 |
|---|---|
| Length | 131 |
| Sequences | 6 |
| Columns | 146 |
| Reading direction | forward |
| Mean pairwise identity | 75.38 |
| Shannon entropy | 0.43753 |
| G+C content | 0.51929 |
| Mean single sequence MFE | -43.07 |
| Consensus MFE | -30.08 |
| Energy contribution | -30.88 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
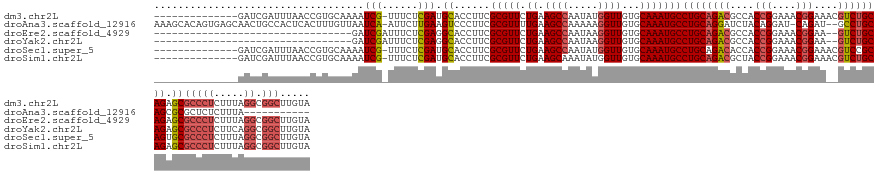
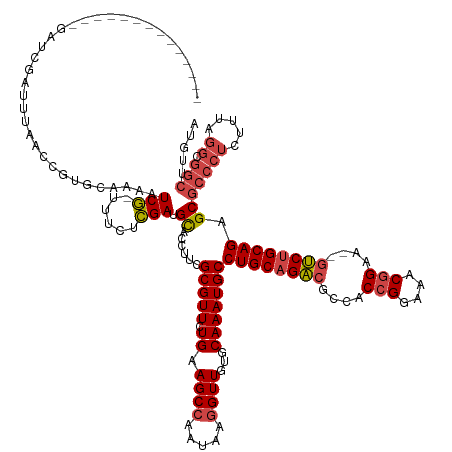
>dm3.chr2L 2966974 131 + 23011544 --------------GAUCGAUUUAACCGUGCAAAAUCG-UUUCUCGAUGCACCUUCGCGUUCUGAAGCCAAUAUGGUUGUGCAAAUGCCUGCAGACGCCACCGGAAACGGAAACGUCUGCAGAGCGCCCUCUUUAGGCGGCUUGUA --------------........((((((((.......(-((((..(((((......)))))..)))))...))))))))(((((..(((((((((((...(((....)))...))))))))).))(((.((....)).)))))))) ( -48.70, z-score = -3.25, R) >droAna3.scaffold_12916 793912 131 + 16180835 AAAGCACAGUGAGCAACUGCCACUCACUUUGUUAAUCA-AUUCUUGAAGUCCCUUCGCGUUUUGAAGCCAAAAAGGUUGUGCAAAUGCCUGCAGGAUCUACAGGAU-CAGAU--GCCUGCAGCGCGCUCUCUUUA----------- ...((((((((((.........)))))).......(((-(..(.(((((...))))).)..))))((((.....))))))))....(((((((((((((.......-.))))--.))))))).))..........----------- ( -39.20, z-score = -1.33, R) >droEre2.scaffold_4929 3012656 111 + 26641161 ---------------------------------GAUCGAUUUCUCGAGGCACCUUCGCGUUCUGAAGCCAAUAAGGUUGUGCAAAUGCCUGCAGACGCCACCGGAAACGGAA--GUCUGCAGAGCGCCCUCUUUAGGCGGCUUGUA ---------------------------------..((((....))))(((......(((((.((.((((.....))))...)))))))((((((((....(((....)))..--))))))))..((((.......))))))).... ( -40.20, z-score = -1.46, R) >droYak2.chr2L 2960267 111 + 22324452 ---------------------------------GAUCGAUUUCUCGAGGCACCUUCGCGUUCUGAAGCCAAUAAGGUUGUGCAAAUGCCUGCAGACGCCACCGGAAACGGAA--GUCUGCAGAGCGCCCUCUUCAGGCGGCUUGUA ---------------------------------..((((....))))(((......(((((.((.((((.....))))...)))))))((((((((....(((....)))..--))))))))..((((.......))))))).... ( -40.20, z-score = -1.31, R) >droSec1.super_5 1122870 131 + 5866729 --------------GAUCGAUUUAACCGUGCAAAAUCG-UUUCUCGAUGCACCUUCGCGUUCUGAAGCCAAUAUGGUUGUGCAAAUGCCUGCAGACACCACCGGAAACGGAAACGUCCGCAGUGCGCCCUCUUUAGGCGGCUUGUA --------------........((((((((.......(-((((..(((((......)))))..)))))...))))))))(((((..((((((.(((....(((....)))....))).)))).))(((.((....)).)))))))) ( -41.40, z-score = -1.45, R) >droSim1.chr2L 2925644 131 + 22036055 --------------GAUCGAUUUAACCGUGCAAAAUCG-UUUCUCGAUGCACCUUCGCGUUCUGAAGCAAAUAUGGUUGUGCAAAUGCCUGCAGACGCUACCGGAAACGGAAACGUCUGCAGAGCGCCCUCUUUAGGCGGCUUGUA --------------........((((((((.......(-((((..(((((......)))))..)))))...))))))))(((((..(((((((((((...(((....)))...))))))))).))(((.((....)).)))))))) ( -48.70, z-score = -3.41, R) >consensus ______________GAUCGAUUUAACCGUGCAAAAUCG_UUUCUCGAUGCACCUUCGCGUUCUGAAGCCAAUAAGGUUGUGCAAAUGCCUGCAGACGCCACCGGAAACGGAA__GUCUGCAGAGCGCCCUCUUUAGGCGGCUUGUA ...................................(((......))).((......(((((.((.((((.....))))...)))))))((((((((....(((....)))....)))))))).))(((.((....)).)))..... (-30.08 = -30.88 + 0.81)
| Location | 2,966,974 – 2,967,105 |
|---|---|
| Length | 131 |
| Sequences | 6 |
| Columns | 146 |
| Reading direction | reverse |
| Mean pairwise identity | 75.38 |
| Shannon entropy | 0.43753 |
| G+C content | 0.51929 |
| Mean single sequence MFE | -44.07 |
| Consensus MFE | -30.12 |
| Energy contribution | -30.77 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
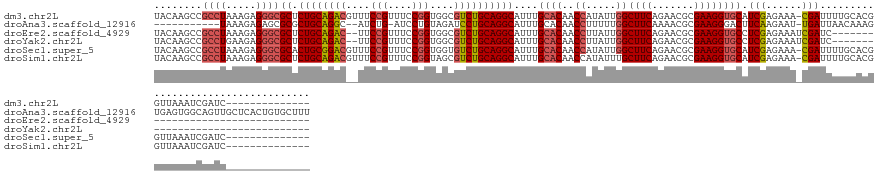
>dm3.chr2L 2966974 131 - 23011544 UACAAGCCGCCUAAAGAGGGCGCUCUGCAGACGUUUCCGUUUCCGGUGGCGUCUGCAGGCAUUUGCACAACCAUAUUGGCUUCAGAACGCGAAGGUGCAUCGAGAAA-CGAUUUUGCACGGUUAAAUCGAUC-------------- .....((.((((.....))))((.((((((((((..(((....)))..))))))))))))....))..((((.......((((.......))))((((((((.....-)))...))))))))).........-------------- ( -48.80, z-score = -2.85, R) >droAna3.scaffold_12916 793912 131 - 16180835 -----------UAAAGAGAGCGCGCUGCAGGC--AUCUG-AUCCUGUAGAUCCUGCAGGCAUUUGCACAACCUUUUUGGCUUCAAAACGCGAAGGGACUUCAAGAAU-UGAUUAACAAAGUGAGUGGCAGUUGCUCACUGUGCUUU -----------.....(((((((.(((((((.--(((((-......))))))))))))(((.((((.((..(.(((((...((((..(..((((...))))..)..)-)))....))))).)..)))))).))).....))))))) ( -41.30, z-score = -0.97, R) >droEre2.scaffold_4929 3012656 111 - 26641161 UACAAGCCGCCUAAAGAGGGCGCUCUGCAGAC--UUCCGUUUCCGGUGGCGUCUGCAGGCAUUUGCACAACCUUAUUGGCUUCAGAACGCGAAGGUGCCUCGAGAAAUCGAUC--------------------------------- ...(((((((((.....))))((.((((((((--..(((....)))....)))))))))).................))))).....(((....)))..((((....))))..--------------------------------- ( -40.70, z-score = -1.79, R) >droYak2.chr2L 2960267 111 - 22324452 UACAAGCCGCCUGAAGAGGGCGCUCUGCAGAC--UUCCGUUUCCGGUGGCGUCUGCAGGCAUUUGCACAACCUUAUUGGCUUCAGAACGCGAAGGUGCCUCGAGAAAUCGAUC--------------------------------- ...(((((((((.....))))((.((((((((--..(((....)))....)))))))))).................))))).....(((....)))..((((....))))..--------------------------------- ( -41.00, z-score = -1.53, R) >droSec1.super_5 1122870 131 - 5866729 UACAAGCCGCCUAAAGAGGGCGCACUGCGGACGUUUCCGUUUCCGGUGGUGUCUGCAGGCAUUUGCACAACCAUAUUGGCUUCAGAACGCGAAGGUGCAUCGAGAAA-CGAUUUUGCACGGUUAAAUCGAUC-------------- .....((.((((.....))))((.((((((((((..(((....)))..))))))))))))....))..((((.......((((.......))))((((((((.....-)))...))))))))).........-------------- ( -43.30, z-score = -1.24, R) >droSim1.chr2L 2925644 131 - 22036055 UACAAGCCGCCUAAAGAGGGCGCUCUGCAGACGUUUCCGUUUCCGGUAGCGUCUGCAGGCAUUUGCACAACCAUAUUUGCUUCAGAACGCGAAGGUGCAUCGAGAAA-CGAUUUUGCACGGUUAAAUCGAUC-------------- .....((.((((.....))))((.(((((((((((.(((....))).)))))))))))))....))..((((...(((((........))))).((((((((.....-)))...))))))))).........-------------- ( -49.30, z-score = -3.66, R) >consensus UACAAGCCGCCUAAAGAGGGCGCUCUGCAGAC__UUCCGUUUCCGGUGGCGUCUGCAGGCAUUUGCACAACCAUAUUGGCUUCAGAACGCGAAGGUGCAUCGAGAAA_CGAUUUUGCACGGUUAAAUCGAUC______________ ........((((.....))))((.((((((((....(((....)))....))))))))))....((((..((.....))((((.......)))))))).(((......)))................................... (-30.12 = -30.77 + 0.64)
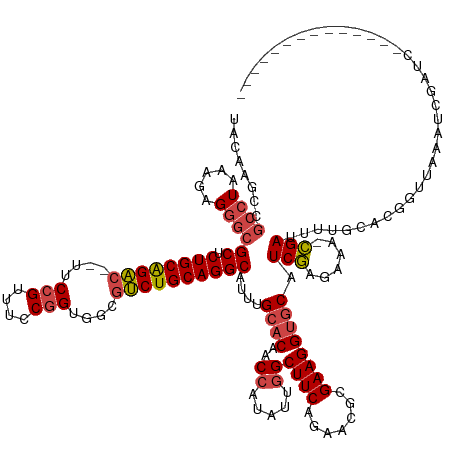
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:42 2011