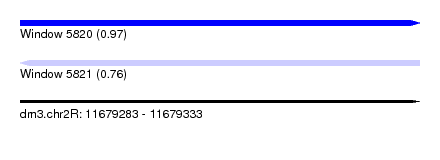
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,679,283 – 11,679,333 |
| Length | 50 |
| Max. P | 0.972570 |

| Location | 11,679,283 – 11,679,333 |
|---|---|
| Length | 50 |
| Sequences | 4 |
| Columns | 52 |
| Reading direction | forward |
| Mean pairwise identity | 87.58 |
| Shannon entropy | 0.19448 |
| G+C content | 0.42596 |
| Mean single sequence MFE | -14.73 |
| Consensus MFE | -12.77 |
| Energy contribution | -12.40 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
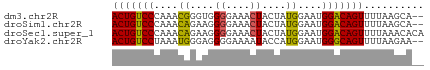
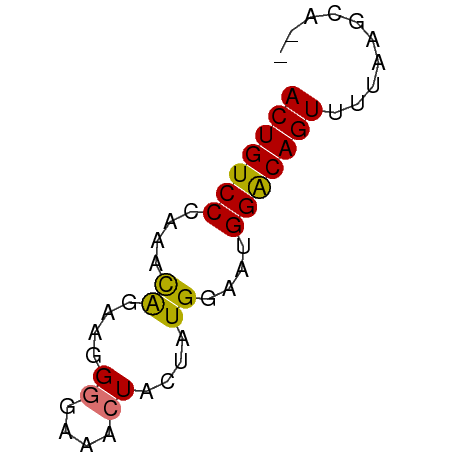
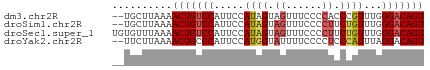
>dm3.chr2R 11679283 50 + 21146708 ACUGUCCCAAACGGGUGGGGAAACUACUAUGGAAUGGACAGUUUUAAGCA-- (((((((......(((((.....))))).......)))))))........-- ( -15.22, z-score = -1.89, R) >droSim1.chr2R 10416409 50 + 19596830 ACUGUCCCAAACAGAAGGGGAAACUACUAUGGAAUGGACAGUUUUAAGCA-- (((((((....((....((....))....))....)))))))........-- ( -15.20, z-score = -2.71, R) >droSec1.super_1 9172875 52 + 14215200 ACUGUCCCAAACAGAAGGGGAAACUACUAUGGAAUGGACAGUUUUAAACACA (((((((....((....((....))....))....))))))).......... ( -15.20, z-score = -2.79, R) >droYak2.chr2R 3831727 50 - 21139217 ACUGUCCUAAAUGGGAGGGGAAAAUACCAUGGAAUGGGCAGUUUUAAGAA-- (((((((...((((............)))).....)))))))........-- ( -13.30, z-score = -1.85, R) >consensus ACUGUCCCAAACAGAAGGGGAAACUACUAUGGAAUGGACAGUUUUAAGCA__ (((((((....((....((....))....))....))))))).......... (-12.77 = -12.40 + -0.37)
| Location | 11,679,283 – 11,679,333 |
|---|---|
| Length | 50 |
| Sequences | 4 |
| Columns | 52 |
| Reading direction | reverse |
| Mean pairwise identity | 87.58 |
| Shannon entropy | 0.19448 |
| G+C content | 0.42596 |
| Mean single sequence MFE | -9.69 |
| Consensus MFE | -8.74 |
| Energy contribution | -9.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.760080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
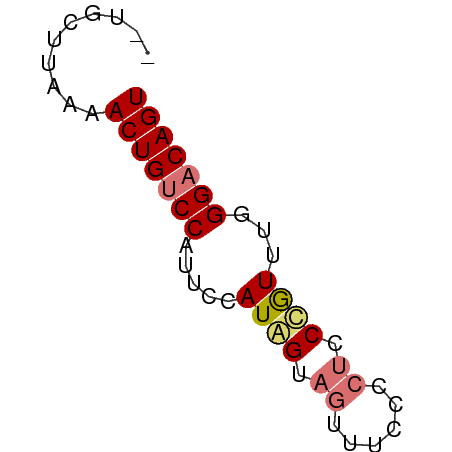
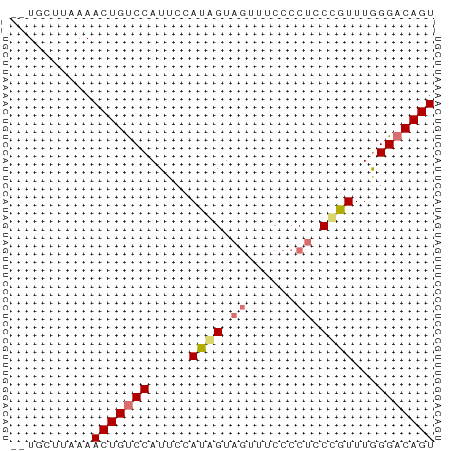
>dm3.chr2R 11679283 50 - 21146708 --UGCUUAAAACUGUCCAUUCCAUAGUAGUUUCCCCACCCGUUUGGGACAGU --........(((((((........((.(.....).)).......))))))) ( -8.26, z-score = -0.55, R) >droSim1.chr2R 10416409 50 - 19596830 --UGCUUAAAACUGUCCAUUCCAUAGUAGUUUCCCCUUCUGUUUGGGACAGU --........(((((((.....((((.((......)).))))...))))))) ( -12.30, z-score = -2.31, R) >droSec1.super_1 9172875 52 - 14215200 UGUGUUUAAAACUGUCCAUUCCAUAGUAGUUUCCCCUUCUGUUUGGGACAGU ..........(((((((.....((((.((......)).))))...))))))) ( -12.30, z-score = -2.29, R) >droYak2.chr2R 3831727 50 + 21139217 --UUCUUAAAACUGCCCAUUCCAUGGUAUUUUCCCCUCCCAUUUAGGACAGU --........((((.((.....((((............))))...)).)))) ( -5.90, z-score = -0.28, R) >consensus __UGCUUAAAACUGUCCAUUCCAUAGUAGUUUCCCCUCCCGUUUGGGACAGU ..........(((((((.....((((.((......)).))))...))))))) ( -8.74 = -9.30 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:31 2011