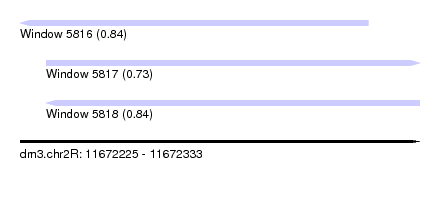
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,672,225 – 11,672,333 |
| Length | 108 |
| Max. P | 0.836776 |

| Location | 11,672,225 – 11,672,319 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.27 |
| Shannon entropy | 0.51404 |
| G+C content | 0.39986 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -13.27 |
| Energy contribution | -13.29 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.836157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
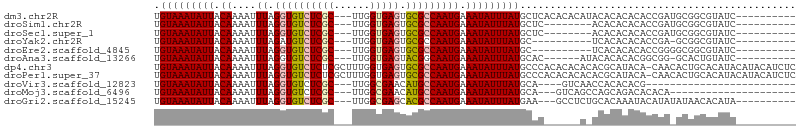
>dm3.chr2R 11672225 94 - 21146708 UGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGUGAGUGCGCCAAUGAAAUAUUUAUGCUCACACACAUACACACACACCGAUGCGGCGUAUC---------- ((((.....)))).......((((..((((---((((((.(((.(.............((((........))))).))).)))))).))))..))))---------- ( -18.21, z-score = 0.53, R) >droSim1.chr2R 10409243 86 - 19596830 UGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGUGAGUGCGCCAAUGAAAUAUUUAUGCUC--------ACACACACACCGAUGCGGCGUAUC---------- ((((.....)))).......((((..((((---((((((.(((.(...((((.....))))....--------.).))).)))))).))))..))))---------- ( -19.10, z-score = -0.22, R) >droSec1.super_1 9165804 86 - 14215200 UGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGUGAGUGCGCCAAUGAAAUAUUUAUGCUC--------ACACACACACCGAUGCGGCGUAUC---------- ((((.....)))).......((((..((((---((((((.(((.(...((((.....))))....--------.).))).)))))).))))..))))---------- ( -19.10, z-score = -0.22, R) >droYak2.chr2R 3824359 83 + 21139217 UGUAAAUAUUACAAAAUUUAGAUGUCUCGC---UUGGUGAGUGCGCCAAUGAAAUAUUUAUGC----------UCACACACACCGA-GCGGCGUAUC---------- ((((.....)))).......((((..((((---((((((.(((.(...((((.....))))..----------.).))).))))))-))))..))))---------- ( -24.80, z-score = -2.98, R) >droEre2.scaffold_4845 8398302 84 + 22589142 UGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGUGAGUGCGCCAAUGAAAUAUUUAUGC----------UCACACACACCGGGGCGGCGUAUC---------- ((((.....)))).......((((..((((---((((((.(((.(...((((.....))))..----------.).))).)))).))))))..))))---------- ( -19.60, z-score = -0.23, R) >droAna3.scaffold_13266 7354936 87 + 19884421 UGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGUGAGUACGGCAAUGAAAUAUUUAUGCAC------AUACACACACGGCGG-GCACUGUAUC---------- ((((.....)))).......((((((.(((---(..(((.(((.((((.(((.....)))))).)------.))).)))..)))))-))))).....---------- ( -22.50, z-score = -2.07, R) >dp4.chr3 266919 106 + 19779522 UGUAAAUAUUACAAAAUUUAGGUGUCUCUCGCUUUGGUGAGUGCGCCAAUGAAAUAUUUAUGCCCACACACACACGCAUACA-CAACACUGCACAUACAUACAUCUC .(((((((((.((....((.(((((..(((((....))))).))))))))).)))))))))..............(((....-......)))............... ( -18.40, z-score = -0.32, R) >droPer1.super_37 245915 106 + 760358 UGUAAAUAUUACAAAAUUUAGGUGUCUCUCGCUUUGGUGAGUGCGCCAAUGAAAUAUUUAUGCCCACACACACACGCAUACA-CAACACUGCACAUACAUACAUCUC .(((((((((.((....((.(((((..(((((....))))).))))))))).)))))))))..............(((....-......)))............... ( -18.40, z-score = -0.32, R) >droVir3.scaffold_12823 1081240 75 + 2474545 UGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGCGAACAUGCCAAUGAAAUAUUUAUGCA----GUCAACCACACACG------------------------- ((((.....)))).....(((((((.(((.---(((((......)))))))).)))))))....----..............------------------------- ( -12.20, z-score = -0.24, R) >droMoj3.scaffold_6496 508932 80 - 26866924 UGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGCGAACAUGCCAAUGAAAUAUUUAUGCA---GUCAGCCAGCAGACACACA--------------------- ((((.....))))........((((((.((---(.(((..((.(((..((((.....)))))))---))..))))))))))))...--------------------- ( -22.30, z-score = -2.84, R) >droGri2.scaffold_15245 5853577 91 - 18325388 UGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGCGAGCACGCCAAUGAAAUAUUUAUGAA---GCCUCUGCACAAAUACAUAUAUAACACAUA---------- .(((((((((.((....((.((((((((((---...))))).))))))))).)))))))))...---((....))......................---------- ( -18.00, z-score = -1.41, R) >consensus UGUAAAUAUUACAAAAUUUAGGUGUCUCGC___UUGGUGAGUGCGCCAAUGAAAUAUUUAUGCAC______A_ACACACACACCGA_GCGGCGUAUC__________ .(((((((((.((....((.((((((((((......))))).))))))))).))))))))).............................................. (-13.27 = -13.29 + 0.02)

| Location | 11,672,232 – 11,672,333 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Shannon entropy | 0.43733 |
| G+C content | 0.37628 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -13.06 |
| Energy contribution | -12.80 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11672232 101 + 21146708 CGCAUCGGUGUGUGUGUAUGUG-UGUGAGCAUAAAUAUUUCAUUGGCGCACUCACCAA---GCGAGACACCUAAAUUUUGUAAUAUUUACAAAGUUGCCAAAAAA- (((...((((.(((((((((.(-(((........))))..)))..)))))).))))..---))).........(((((((((.....))))))))).........- ( -25.70, z-score = -1.21, R) >droSim1.chr2R 10409250 94 + 19596830 --------CGCAUCGGUGUGUG-UGUGAGCAUAAAUAUUUCAUUGGCGCACUCACCAA---GCGAGACACCUAAAUUUUGUAAUAUUUACAAAGUUGCCAAAAAAA --------(((...((((.(((-(((.((.............)).)))))).))))..---))).........(((((((((.....))))))))).......... ( -23.02, z-score = -1.57, R) >droSec1.super_1 9165811 94 + 14215200 --------CGCAUCGGUGUGUG-UGUGAGCAUAAAUAUUUCAUUGGCGCACUCACCAA---GCGAGACACCUAAAUUUUGUAAUAUUUACAAAGUUGCCAAAAAAA --------(((...((((.(((-(((.((.............)).)))))).))))..---))).........(((((((((.....))))))))).......... ( -23.02, z-score = -1.57, R) >droYak2.chr2R 3824366 90 - 21139217 ----------CGCUC-GGUGUG-UGUGAGCAUAAAUAUUUCAUUGGCGCACUCACCAA---GCGAGACAUCUAAAUUUUGUAAUAUUUACAAAGUUGCCAAAAAA- ----------((((.-((((.(-((((..((............)).))))).)))).)---))).........(((((((((.....))))))))).........- ( -25.20, z-score = -2.98, R) >droEre2.scaffold_4845 8398309 91 - 22589142 ----------CGCCCCGGUGUG-UGUGAGCAUAAAUAUUUCAUUGGCGCACUCACCAA---GCGAGACACCUAAAUUUUGUAAUAUUUACAAAGUUGCCAAAAAA- ----------(((...((((.(-((((..((............)).))))).))))..---))).........(((((((((.....))))))))).........- ( -21.90, z-score = -1.76, R) >droAna3.scaffold_13266 7354943 91 - 19884421 ------UGCCCGC-CGUGUGUG-UAUGUGCAUAAAUAUUUCAUUGCCGUACUCACCAA---GCGAGACACCUAAAUUUUGUAAUAUUUACAAAGUUGCCAAA---- ------.((.(((-...(((.(-((((.(((............)))))))).)))...---))).........(((((((((.....)))))))))))....---- ( -21.40, z-score = -2.36, R) >dp4.chr3 266935 103 - 19779522 CAGUGUUGUGUAUGCGUGUGUG-UGUGGGCAUAAAUAUUUCAUUGGCGCACUCACCAAAGCGAGAGACACCUAAAUUUUGUAAUAUUUACAAAGUUGCCAAAAA-- ..(((((...(.(((.((.(((-.(((.((...(((.....))).)).))).)))))..))).).)))))...(((((((((.....)))))))))........-- ( -24.10, z-score = -0.12, R) >droPer1.super_37 245931 103 - 760358 CAGUGUUGUGUAUGCGUGUGUG-UGUGGGCAUAAAUAUUUCAUUGGCGCACUCACCAAAGCGAGAGACACCUAAAUUUUGUAAUAUUUACAAAGUUGCCAAAAA-- ..(((((...(.(((.((.(((-.(((.((...(((.....))).)).))).)))))..))).).)))))...(((((((((.....)))))))))........-- ( -24.10, z-score = -0.12, R) >droVir3.scaffold_12823 1081246 84 - 2474545 -------------------UGGUUGACUGCAUAAAUAUUUCAUUGGCAUGUUCGCCAA---GCGAGACACCUAAAUUUUGUAAUAUUUACAAAGUUGCUAAAAAUG -------------------..(((....((.......((((.(((((......)))))---..))))......(((((((((.....)))))))))))....))). ( -16.70, z-score = -1.16, R) >droMoj3.scaffold_6496 508939 88 + 26866924 ---------------CUGCUGGCUGACUGCAUAAAUAUUUCAUUGGCAUGUUCGCCAA---GCGAGACACCUAAAUUUUGUAAUAUUUACAAAGUUGCUAAAAAUG ---------------.(((((((.((((((...(((.....))).))).))).))).)---))).........(((((((((.....))))))))).......... ( -19.50, z-score = -1.55, R) >droGri2.scaffold_15245 5853584 99 + 18325388 ----UAUAUAUGUAUUUGUGCAGAGGCUUCAUAAAUAUUUCAUUGGCGUGCUCGCCAA---GCGAGACACCUAAAUUUUGUAAUAUUUACAAAGUUGCUAAAAAUG ----.......(((((((((.((...)).)))))))))....(((((((((((((...---))))).)))...(((((((((.....))))))))))))))..... ( -25.40, z-score = -2.64, R) >consensus _________GCGU_CGUGUGUG_UGUGAGCAUAAAUAUUUCAUUGGCGCACUCACCAA___GCGAGACACCUAAAUUUUGUAAUAUUUACAAAGUUGCCAAAAAA_ ..........................................(((((...(((.(......).))).......(((((((((.....))))))))))))))..... (-13.06 = -12.80 + -0.26)



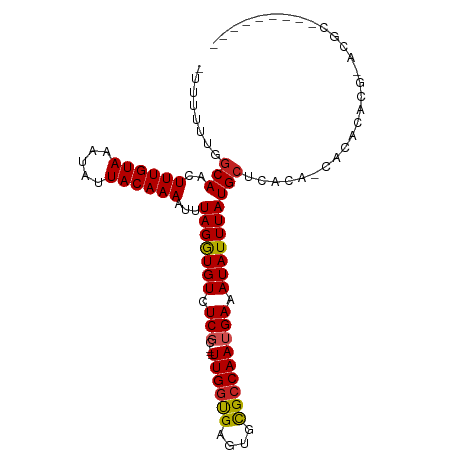
| Location | 11,672,232 – 11,672,333 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.20 |
| Shannon entropy | 0.43733 |
| G+C content | 0.37628 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -15.02 |
| Energy contribution | -15.13 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
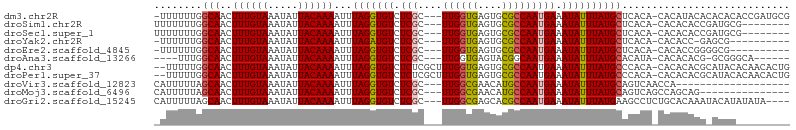
>dm3.chr2R 11672232 101 - 21146708 -UUUUUUGGCAACUUUGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGUGAGUGCGCCAAUGAAAUAUUUAUGCUCACA-CACAUACACACACACCGAUGCG -.......(((..((((((.....)))))).....(((((.....---...(((.(((.((..((((.....)))))).))).-)))........)))))..))). ( -21.19, z-score = -0.85, R) >droSim1.chr2R 10409250 94 - 19596830 UUUUUUUGGCAACUUUGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGUGAGUGCGCCAAUGAAAUAUUUAUGCUCACA-CACACACCGAUGCG-------- ........(((..((((((.....)))))).....((((((....---..)(((.(((.((..((((.....)))))).))).-))))))))..))).-------- ( -18.80, z-score = -0.28, R) >droSec1.super_1 9165811 94 - 14215200 UUUUUUUGGCAACUUUGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGUGAGUGCGCCAAUGAAAUAUUUAUGCUCACA-CACACACCGAUGCG-------- ........(((..((((((.....)))))).....((((((....---..)(((.(((.((..((((.....)))))).))).-))))))))..))).-------- ( -18.80, z-score = -0.28, R) >droYak2.chr2R 3824366 90 + 21139217 -UUUUUUGGCAACUUUGUAAAUAUUACAAAAUUUAGAUGUCUCGC---UUGGUGAGUGCGCCAAUGAAAUAUUUAUGCUCACA-CACACC-GAGCG---------- -......((((.(((((((.....))))).....)).)))).(((---((((((.(((.(...((((.....))))...).))-).))))-)))))---------- ( -23.60, z-score = -2.68, R) >droEre2.scaffold_4845 8398309 91 + 22589142 -UUUUUUGGCAACUUUGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGUGAGUGCGCCAAUGAAAUAUUUAUGCUCACA-CACACCGGGGCG---------- -(((.(((((...((((((.....))))))......(((.(((((---...))))))))))))).)))........((((.(.-......))))).---------- ( -19.50, z-score = -0.24, R) >droAna3.scaffold_13266 7354943 91 + 19884421 ----UUUGGCAACUUUGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGUGAGUACGGCAAUGAAAUAUUUAUGCACAUA-CACACACG-GCGGGCA------ ----...(....)((((((.....)))))).......((((.(((---(..(((.(((.((((.(((.....)))))).).))-).)))..)-)))))))------ ( -19.80, z-score = -0.88, R) >dp4.chr3 266935 103 + 19779522 --UUUUUGGCAACUUUGUAAAUAUUACAAAAUUUAGGUGUCUCUCGCUUUGGUGAGUGCGCCAAUGAAAUAUUUAUGCCCACA-CACACACGCAUACACAACACUG --.....((((..((((((.....)))))).....(((((..(((((....))))).))))).............))))....-...................... ( -22.20, z-score = -1.35, R) >droPer1.super_37 245931 103 + 760358 --UUUUUGGCAACUUUGUAAAUAUUACAAAAUUUAGGUGUCUCUCGCUUUGGUGAGUGCGCCAAUGAAAUAUUUAUGCCCACA-CACACACGCAUACACAACACUG --.....((((..((((((.....)))))).....(((((..(((((....))))).))))).............))))....-...................... ( -22.20, z-score = -1.35, R) >droVir3.scaffold_12823 1081246 84 + 2474545 CAUUUUUAGCAACUUUGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGCGAACAUGCCAAUGAAAUAUUUAUGCAGUCAACCA------------------- ........(((..((((((.....))))))...(((((((.(((.---(((((......)))))))).)))))))))).........------------------- ( -17.50, z-score = -1.64, R) >droMoj3.scaffold_6496 508939 88 - 26866924 CAUUUUUAGCAACUUUGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGCGAACAUGCCAAUGAAAUAUUUAUGCAGUCAGCCAGCAG--------------- ........(((..((((((.....))))))...(((((((.(((.---(((((......)))))))).)))))))))).............--------------- ( -17.50, z-score = -0.65, R) >droGri2.scaffold_15245 5853584 99 - 18325388 CAUUUUUAGCAACUUUGUAAAUAUUACAAAAUUUAGGUGUCUCGC---UUGGCGAGCACGCCAAUGAAAUAUUUAUGAAGCCUCUGCACAAAUACAUAUAUA---- ........(((.(((..((((((((.((....((.((((((((((---...))))).))))))))).))))))))..)))....)))...............---- ( -24.60, z-score = -3.04, R) >consensus _UUUUUUGGCAACUUUGUAAAUAUUACAAAAUUUAGGUGUCUCGC___UUGGUGAGUGCGCCAAUGAAAUAUUUAUGCUCACA_CACACACG_ACGC_________ ........(((..((((((.....)))))).....((((((((((......))))).))))).............)))............................ (-15.02 = -15.13 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:29 2011