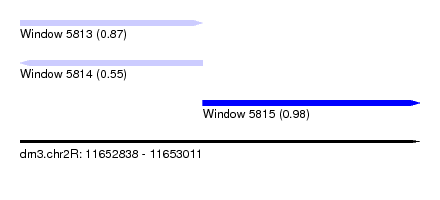
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,652,838 – 11,653,011 |
| Length | 173 |
| Max. P | 0.979599 |

| Location | 11,652,838 – 11,652,917 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 83.00 |
| Shannon entropy | 0.32245 |
| G+C content | 0.51860 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -19.23 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.867143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
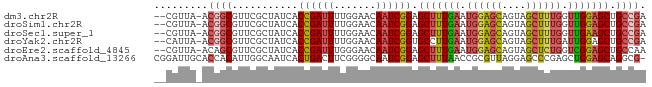
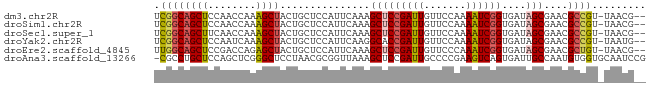
>dm3.chr2R 11652838 79 + 21146708 --CGUUA-ACGGCGUUCGCUAUCACCGAUUUUGGAACAAUCGGAGCUUUGAAUGGAGCAGUAGCUUUGGUUGGAGCUGCCGA --.....-.((((...........((((((.......))))))(((((..(..(((((....)))))..)..))))))))). ( -27.50, z-score = -2.04, R) >droSim1.chr2R 10388668 79 + 19596830 --CGUUA-ACGGCGUUCGCUAUCACCGAUUUUGGAACAAUCGGAGCUUUGAAUGGAGCAGUAGCUUUGGUUGGAGCUGCCGA --.....-.((((...........((((((.......))))))(((((..(..(((((....)))))..)..))))))))). ( -27.50, z-score = -2.04, R) >droSec1.super_1 9146422 79 + 14215200 --CGUUA-ACGGCGUUCGCUAUCACCGAUUUUGGAACAAUCGGAGCUUUGAAUGGAGCAGUAGCUUUGGUUGAAGCUGCCGA --.....-.((((...........((((((.......))))))(((((..(..(((((....)))))..)..))))))))). ( -27.10, z-score = -2.09, R) >droYak2.chr2R 3804667 79 - 21139217 --CAUUA-ACGGCGUUCGCUAUCACCGAUUUUGGAACAAUCGGUGCCUUGAAUGGAGCAGUAGCUUUGAUUGGAGCUGCCGA --.....-.((((....(((..((((((((.......))))))))((......)))))...((((((....)))))))))). ( -26.50, z-score = -2.10, R) >droEre2.scaffold_4845 8379226 79 - 22589142 --CGUUA-ACAGCGUUCGCUAUCACCGAUUUGGGAACAAUCGGAGCUUUGAAUGGAGCAGUAGCUCUGGUCGGAGCUGCCAA --.....-..(((....)))....((((((.(....)))))))((((((((..(((((....)))))..))))))))..... ( -28.60, z-score = -2.23, R) >droAna3.scaffold_13266 7336975 81 - 19884421 CGGAUUGCACCACAUUGGCAAUCACUGACUUCGGGGCAAUCGGAGCUUUAACCGCGUUAGGAGCCCGAGCUGGAGCAGGCG- ..((((((.........)))))).(((.((((((.....((((.((((((((...)))).)))))))).)))))))))...- ( -26.90, z-score = -0.92, R) >consensus __CGUUA_ACGGCGUUCGCUAUCACCGAUUUUGGAACAAUCGGAGCUUUGAAUGGAGCAGUAGCUUUGGUUGGAGCUGCCGA .........((((...........((((((.......)))))).(((((((.((((((....)))))).))))))).)))). (-19.23 = -19.98 + 0.76)
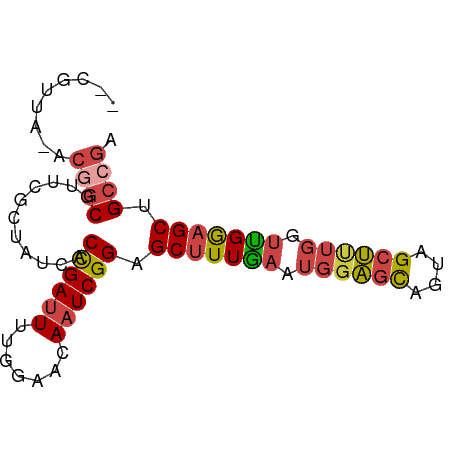
| Location | 11,652,838 – 11,652,917 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 83.00 |
| Shannon entropy | 0.32245 |
| G+C content | 0.51860 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -14.02 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
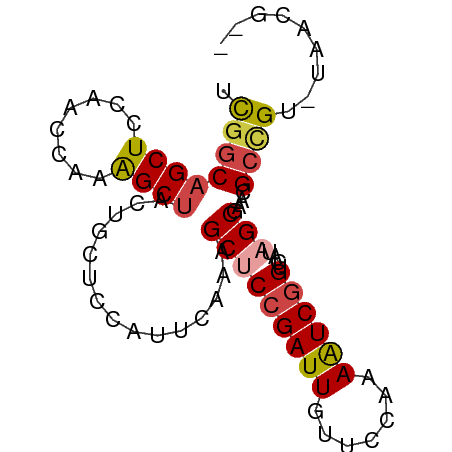
>dm3.chr2R 11652838 79 - 21146708 UCGGCAGCUCCAACCAAAGCUACUGCUCCAUUCAAAGCUCCGAUUGUUCCAAAAUCGGUGAUAGCGAACGCCGU-UAACG-- .((((((((........))))...............(((((((((.......))))))....)))....)))).-.....-- ( -18.80, z-score = -1.64, R) >droSim1.chr2R 10388668 79 - 19596830 UCGGCAGCUCCAACCAAAGCUACUGCUCCAUUCAAAGCUCCGAUUGUUCCAAAAUCGGUGAUAGCGAACGCCGU-UAACG-- .((((((((........))))...............(((((((((.......))))))....)))....)))).-.....-- ( -18.80, z-score = -1.64, R) >droSec1.super_1 9146422 79 - 14215200 UCGGCAGCUUCAACCAAAGCUACUGCUCCAUUCAAAGCUCCGAUUGUUCCAAAAUCGGUGAUAGCGAACGCCGU-UAACG-- .(((((((((......)))))...............(((((((((.......))))))....)))....)))).-.....-- ( -19.90, z-score = -1.96, R) >droYak2.chr2R 3804667 79 + 21139217 UCGGCAGCUCCAAUCAAAGCUACUGCUCCAUUCAAGGCACCGAUUGUUCCAAAAUCGGUGAUAGCGAACGCCGU-UAAUG-- .((((((((........))))..((((((......))((((((((.......))))))))..))))...)))).-.....-- ( -23.50, z-score = -2.80, R) >droEre2.scaffold_4845 8379226 79 + 22589142 UUGGCAGCUCCGACCAGAGCUACUGCUCCAUUCAAAGCUCCGAUUGUUCCCAAAUCGGUGAUAGCGAACGCUGU-UAACG-- ((((((((...((...((((....))))...))...(((((((((.......))))))....)))....)))))-)))..-- ( -24.30, z-score = -2.58, R) >droAna3.scaffold_13266 7336975 81 + 19884421 -CGCCUGCUCCAGCUCGGGCUCCUAACGCGGUUAAAGCUCCGAUUGCCCCGAAGUCAGUGAUUGCCAAUGUGGUGCAAUCCG -..........((((..((((.(....).))))..))))..((((((.(((......((....)).....))).)))))).. ( -18.10, z-score = 1.47, R) >consensus UCGGCAGCUCCAACCAAAGCUACUGCUCCAUUCAAAGCUCCGAUUGUUCCAAAAUCGGUGAUAGCGAACGCCGU_UAACG__ .((((((((........))))...............(((((((((.......))))))....)))....))))......... (-14.02 = -14.50 + 0.48)
| Location | 11,652,917 – 11,653,011 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 91.24 |
| Shannon entropy | 0.14235 |
| G+C content | 0.57678 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -35.10 |
| Energy contribution | -35.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
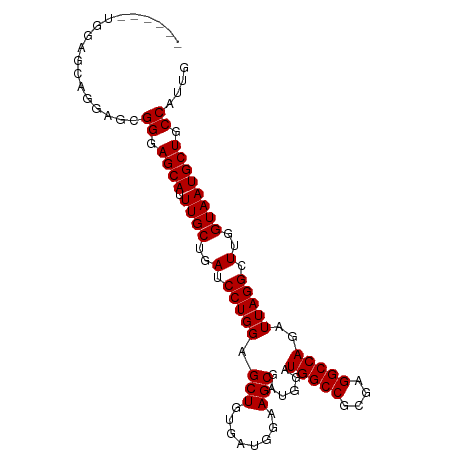
>dm3.chr2R 11652917 94 + 21146708 ------UGGAACAGGAGCGGGAGCACUUGCUGAUCCUGGAGCUGUGAUGGAAGCGAUGGAUGGCCGCGAGGCCAGAUUAGGCUUGGUAAUGCUGCCAUUG ------............((.((((.((((..(.(((((.(((........)))......(((((....)))))..))))).)..)))))))).)).... ( -35.90, z-score = -2.19, R) >droSec1.super_1 9146501 88 + 14215200 ------------UGGAGCGGGAGCAUUUGCUGAUCCUGGAGCUGUGAUGGAAGCGAUGGAUGGCCGCGAGGCCAGAUUAGGCUUGGUAAUGCUGCCAUUG ------------......((.((((((.((..(.(((((.(((........)))......(((((....)))))..))))).)..)))))))).)).... ( -35.90, z-score = -2.78, R) >droYak2.chr2R 3804752 94 - 21139217 ------AGGAGCAGGAGCGGGAGCAUUUGCUGAUCCUGGAGCUGUGAUGGAAGCGAUGGAUGGCCGCGAGGCCAGAUUAGGCUUGGUAAUGCUGCCAUUG ------....((....))((.((((((.((..(.(((((.(((........)))......(((((....)))))..))))).)..)))))))).)).... ( -37.10, z-score = -2.23, R) >droEre2.scaffold_4845 8379311 100 - 22589142 UGGAGCUGCAGCAGGAGCGGGAGCAUUUGCUGAUCCUGGAGCUGUGAUGGAAGCGAUGGAUGGCCGCGAGGCCAGAUUAGGCUGGGUAAUGCUGCCAUUG ....(((.(....).)))((.((((((.(((.(.(((((.(((........)))......(((((....)))))..))))).).))))))))).)).... ( -39.20, z-score = -1.44, R) >consensus ______UGGAGCAGGAGCGGGAGCAUUUGCUGAUCCUGGAGCUGUGAUGGAAGCGAUGGAUGGCCGCGAGGCCAGAUUAGGCUUGGUAAUGCUGCCAUUG ..................((.((((.((((..(.(((((.(((........)))......(((((....)))))..))))).)..)))))))).)).... (-35.10 = -35.10 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:27 2011