| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,641,180 – 11,641,277 |
| Length | 97 |
| Max. P | 0.971574 |

| Location | 11,641,180 – 11,641,277 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.79 |
| Shannon entropy | 0.53060 |
| G+C content | 0.39683 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -12.42 |
| Energy contribution | -14.13 |
| Covariance contribution | 1.71 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
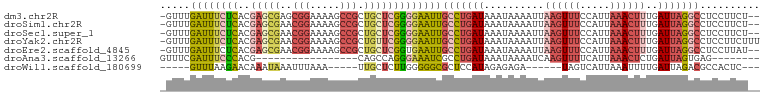
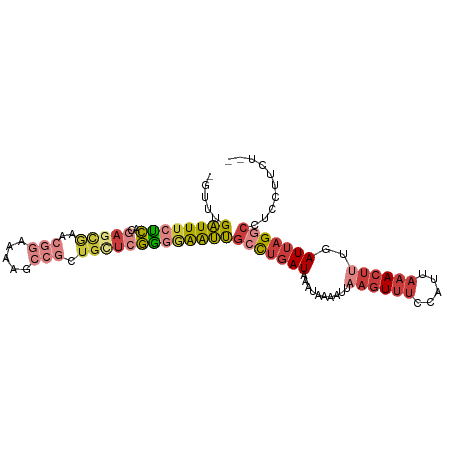
>dm3.chr2R 11641180 97 + 21146708 -GUUUGAUUUCUCACGAGCGAGCGGAAAAGCCGCUGCUCGGGGAAUUGCCUGAUAAAUAAAAUUAAGUUUCCAUUAAACUUUGAUUAGGCCUCCUUCU-- -.......(((((.(((((.(((((.....)))))))))))))))..(((((((..........((((((.....))))))..)))))))........-- ( -31.30, z-score = -3.06, R) >droSim1.chr2R 10376882 97 + 19596830 -GUUUGAUUUCUCACGAGCGAACGGAAAAGCCGCUGCUCGGGGAAUUGCCUGAUAAAUAAAAUUAAGUUUCCAUUAAACUUUGAUUAGGCCUCCUUCU-- -.......(((((.((((((..(((.....))).)))))))))))..(((((((..........((((((.....))))))..)))))))........-- ( -25.80, z-score = -1.85, R) >droSec1.super_1 9134693 97 + 14215200 -GUUUGAUUUCUCACGAGCGAACGGAAAAGCCGCUGCUCGGGGAAUUGCCUGAUAAAUAAAAUUAAGUUUCCAUUAAACUUUGAUUAGGCCUCCUUCU-- -.......(((((.((((((..(((.....))).)))))))))))..(((((((..........((((((.....))))))..)))))))........-- ( -25.80, z-score = -1.85, R) >droYak2.chr2R 3792603 99 - 21139217 -GUUUGAUUUCUCACGAGCGAACGGAAAAGCCGCUGUUCGGGGAAUUGCCUGAUAAAUAAAAUUAAGUUUCCAUUAAACUUUGAUUAGGCCUCCUUCUUU -.......(((((.((((((..(((.....))).)))))))))))..(((((((..........((((((.....))))))..))))))).......... ( -23.40, z-score = -1.08, R) >droEre2.scaffold_4845 8367348 97 - 22589142 -GUUUGAUUUCUCACGAGCGAACGGAAAAGCCGCUGCUCGGUGAAUUGCCUGAUAAAUAAAAUUAAGUUUCCAUUAAACUUUGAUUAGGCCUCCUUAU-- -.......(((.(.((((((..(((.....))).))))))).)))..(((((((..........((((((.....))))))..)))))))........-- ( -22.50, z-score = -1.65, R) >droAna3.scaffold_13266 7325707 75 - 19884421 GUUUCGAUUUCCCACG-----------------CAGCCAGGGAAAUCGCCUGAUAAAUAAAAUCAAGUUUUCAUUAAACUCUGAUUAGUGAG-------- ....(((((((((...-----------------......)))))))))............((((((((((.....))))).)))))......-------- ( -17.70, z-score = -2.61, R) >droWil1.scaffold_180699 1636922 81 - 2593675 -----GUUUAAGAACAAAUAAAUUUAAA-----UUGCUCUUGGGGGCGCUCCAUAGAGAGA------UAGUCAUUAAAUUUUGAUUAGACGCCACUC--- -----.(((((((.(((...........-----))).)))))))(((((((......))).------((((((........))))))..))))....--- ( -15.90, z-score = -1.06, R) >consensus _GUUUGAUUUCUCACGAGCGAACGGAAAAGCCGCUGCUCGGGGAAUUGCCUGAUAAAUAAAAUUAAGUUUCCAUUAAACUUUGAUUAGGCCUCCUUCU__ .....((((((((..((((...(((.....)))..))))))))))))(((((((..........((((((.....))))))..))))))).......... (-12.42 = -14.13 + 1.71)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:23 2011