| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,622,307 – 11,622,406 |
| Length | 99 |
| Max. P | 0.709081 |

| Location | 11,622,307 – 11,622,406 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 71.91 |
| Shannon entropy | 0.61626 |
| G+C content | 0.43399 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -10.90 |
| Energy contribution | -10.58 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.526085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
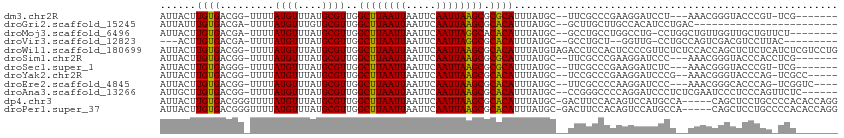
>dm3.chr2R 11622307 99 + 21146708 AUUACUUGUGACGG-UUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAAGCGCGCAUUUAUGC--UUCGCCCGAAGGAUCCU---AAACGGGUACCCGU-UCG------- .........(((((-......((..((((((..((((((((.....))))))))))))))..))..--...(((((.((....))---...)))))..))))-)..------- ( -27.50, z-score = -1.80, R) >droGri2.scaffold_15245 5791384 86 + 18325388 AUUAUUUGUGACGA-UUUUAUGUUUGUGCGUUGGCUUAAUUAAUUCAAUUAAGCGCACAUUUAUGC--GCUUGCUUGCCACAUCCUGAC------------------------ ......((((....-...((((..(((((....((((((((.....)))))))))))))..)))).--((......)))))).......------------------------ ( -18.80, z-score = -1.03, R) >droMoj3.scaffold_6496 437850 101 + 26866924 AUUACUUGUGACGA-UUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAGGCACACAUUUAUGC--GCCUGCCUGGCCUG-CCUGGCUGUUGGUUGCUGUUCU-------- ......((((....-....((((....))))..((((((((.....)))))))).)))).....((--(((.(((.((....-)).)))....)).)))......-------- ( -23.20, z-score = 0.08, R) >droVir3.scaffold_12823 1003279 95 - 2474545 ---ACUUGUGACGA-UUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAGGCGCACAUUUAUGC--GCCUGCU--GGUUG-CCUGCCAGUCGACGUCCUUAC--------- ---...........-............((((((((..............((((((((......)))--))))).(--(((..-...))))))))))))......--------- ( -26.50, z-score = -1.79, R) >droWil1.scaffold_180699 1606162 112 - 2593675 AUUACUUGUGACGG-UUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAAGCGCACAUUUAUGUAGACCUCCACUCCCCGUUCUCUCCACCAGCUCUCUCAUCUCGUCCUG .......(((..((-(((.((((..(((.((..((((((((.....)))))))))).)))..)))))))))..)))..................................... ( -18.80, z-score = -1.70, R) >droSim1.chr2R 10357911 100 + 19596830 AUUACUUGUGACGG-UUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAAGCGCGCAUUUAUGC--UUCGCCCGAAGGAUCCC---AAACGGGUACCCACCUCG------- ..(((((((...((-..((..((..((((((..((((((((.....))))))))))))))..)).(--(((....))))))..))---..))))))).........------- ( -26.30, z-score = -1.53, R) >droSec1.super_1 9115408 99 + 14215200 AUUACUUGUGAGGG-UUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAAGCGCGCAUUUAUGC--UUCGCCCGAAGGAUCUC---AAACGGGUACCCGU-UCG------- ........(((((.-......((..((((((..((((((((.....))))))))))))))..)).(--(((....))))..))))---)(((((....))))-)..------- ( -28.10, z-score = -1.90, R) >droYak2.chr2R 3773304 102 - 21139217 AUUACUUGUGACGG-UUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAAGCGCACAUUUAUGC--UCCGCCCGAAGGAUCCCG--AAACGGGUACCCAG-UCGCC----- .......(((((((-...((((...(((.((..((((((((.....)))))))))).))).)))).--.))(((((..((...)).--...))))).....)-)))).----- ( -25.80, z-score = -0.85, R) >droEre2.scaffold_4845 8348653 102 - 22589142 AUUACUUGUGACGG-UUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAAGCGCACAUUUAUGC--UUCGCCCCAAGGAUCCC---AAACGGGCACCCAG-UCGGUC---- ........((((((-............((((((((((((((.....)))))))).)).....))))--...((((...((...))---....))))..)).)-)))...---- ( -20.90, z-score = 0.50, R) >droAna3.scaffold_13266 7309180 104 - 19884421 AUUGCUUGUGACGG-UUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAAGCGCACAUUUAUGC--CCGGGCCCCAGGAUCCCUCUCGAAUCCCUCCCAGUUCUC------ ...((....((((.-.....))))...))....((((((((.....))))))))(((......)))--..(((.....((((.(.....).))))..))).......------ ( -17.70, z-score = 1.25, R) >dp4.chr3 212837 107 - 19779522 AUUACUUGUGACGGGUUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAAGCGCACAUUUAUGC-GACUUCCACAGUCCAUGCCA-----CAGCUCCUGCCCCACACCAGG ....((.(((..((((.....(((...((((.(((((((((.....))))))))(((......)))-((((.....)))))))))..-----.)))....))))..))).)). ( -24.80, z-score = -1.11, R) >droPer1.super_37 192063 107 - 760358 AUUACUUGUGACGGGUUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAAGCGCACAUUUAUGC-GACUUCCACAGUCCAUGCCA-----CAGCUCCUGCCCCACACCAGG ....((.(((..((((.....(((...((((.(((((((((.....))))))))(((......)))-((((.....)))))))))..-----.)))....))))..))).)). ( -24.80, z-score = -1.11, R) >consensus AUUACUUGUGACGG_UUUUAUGUUUAUGCGUUGGCUUAAUUAAUUCAAUUAAGCGCACAUUUAUGC__UCCGCCCCAAGGAUCCCU__AAACGGGCACCCGC_UCG_______ ......((((.........((((....))))..((((((((.....)))))))).))))...................................................... (-10.90 = -10.58 + -0.33)
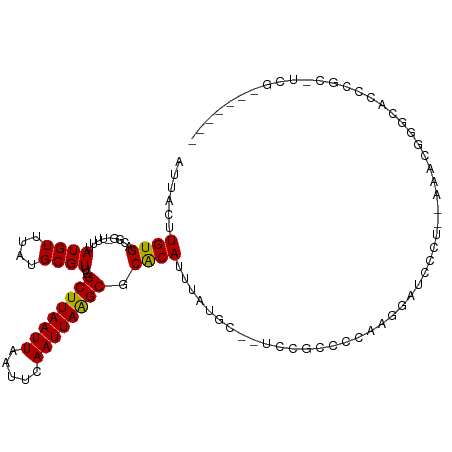

| Location | 11,622,307 – 11,622,406 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 71.91 |
| Shannon entropy | 0.61626 |
| G+C content | 0.43399 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -10.04 |
| Energy contribution | -10.10 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
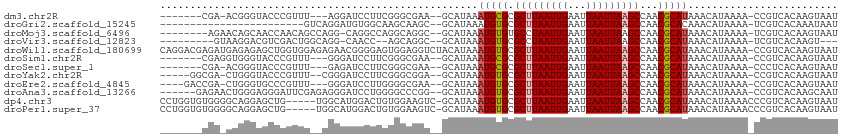
>dm3.chr2R 11622307 99 - 21146708 -------CGA-ACGGGUACCCGUUU---AGGAUCCUUCGGGCGAA--GCAUAAAUGCGCGCUUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAA-CCGUCACAAGUAAU -------.((-((((....))))))---.((..((....))....--......(((((.(((((((((...)))))))))...)))))..........-))............ ( -25.30, z-score = -1.93, R) >droGri2.scaffold_15245 5791384 86 - 18325388 ------------------------GUCAGGAUGUGGCAAGCAAGC--GCAUAAAUGUGCGCUUAAUUGAAUUAAUUAAGCCAACGCACAAACAUAAAA-UCGUCACAAAUAAU ------------------------.......((((((..((....--)).....((((((((((((((...)))))))))....))))).........-..))))))...... ( -21.80, z-score = -2.69, R) >droMoj3.scaffold_6496 437850 101 - 26866924 --------AGAACAGCAACCAACAGCCAGG-CAGGCCAGGCAGGC--GCAUAAAUGUGUGCCUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAA-UCGUCACAAGUAAU --------......((........(((...-..)))..(((((((--((((....))))))))..((((.....)))))))...))............-.............. ( -22.30, z-score = -1.22, R) >droVir3.scaffold_12823 1003279 95 + 2474545 ---------GUAAGGACGUCGACUGGCAGG-CAACC--AGCAGGC--GCAUAAAUGUGCGCCUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAA-UCGUCACAAGU--- ---------.....((((((((((((....-...))--)).((((--((((....))))))))..)))).........((....))............-.))))......--- ( -23.80, z-score = -1.50, R) >droWil1.scaffold_180699 1606162 112 + 2593675 CAGGACGAGAUGAGAGAGCUGGUGGAGAGAACGGGGAGUGGAGGUCUACAUAAAUGUGCGCUUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAA-CCGUCACAAGUAAU .................(((.(((......((((...((((....))))....(((((.(((((((((...)))))))))...)))))..........-))))))).)))... ( -22.50, z-score = -0.89, R) >droSim1.chr2R 10357911 100 - 19596830 -------CGAGGUGGGUACCCGUUU---GGGAUCCUUCGGGCGAA--GCAUAAAUGCGCGCUUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAA-CCGUCACAAGUAAU -------.(((((((((.(((....---))))))).....(((..--(((....)))(.(((((((((...))))))))))..)))...........)-)).))......... ( -25.70, z-score = -1.02, R) >droSec1.super_1 9115408 99 - 14215200 -------CGA-ACGGGUACCCGUUU---GAGAUCCUUCGGGCGAA--GCAUAAAUGCGCGCUUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAA-CCCUCACAAGUAAU -------.((-((((....))))))---(((..((....))....--......(((((.(((((((((...)))))))))...)))))..........-..)))......... ( -26.10, z-score = -2.64, R) >droYak2.chr2R 3773304 102 + 21139217 -----GGCGA-CUGGGUACCCGUUU--CGGGAUCCUUCGGGCGGA--GCAUAAAUGUGCGCUUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAA-CCGUCACAAGUAAU -----..(((-..((((.((((...--)))))))).)))(((((.--((((....))))(((((((((...)))))))))..................-)))))......... ( -30.90, z-score = -2.26, R) >droEre2.scaffold_4845 8348653 102 + 22589142 ----GACCGA-CUGGGUGCCCGUUU---GGGAUCCUUGGGGCGAA--GCAUAAAUGUGCGCUUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAA-CCGUCACAAGUAAU ----..((((-.(((....))).))---))....((((.((((..--((((....))))(((((((((...)))))))))..................-.)))).)))).... ( -27.10, z-score = -1.15, R) >droAna3.scaffold_13266 7309180 104 + 19884421 ------GAGAACUGGGAGGGAUUCGAGAGGGAUCCUGGGGCCCGG--GCAUAAAUGUGCGCUUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAA-CCGUCACAAGCAAU ------.....(((((.(((((((.....)))))))....)))))--((....(((((.(((((((((...)))))))))...)))))..((......-..)).....))... ( -28.50, z-score = -1.55, R) >dp4.chr3 212837 107 + 19779522 CCUGGUGUGGGGCAGGAGCUG-----UGGCAUGGACUGUGGAAGUC-GCAUAAAUGUGCGCUUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAACCCGUCACAAGUAAU ((((........)))).((((-----(((((((((((.....))))-.)))..(((((.(((((((((...)))))))))...))))).............))))).)))... ( -29.50, z-score = -0.82, R) >droPer1.super_37 192063 107 + 760358 CCUGGUGUGGGGCAGGAGCUG-----UGGCAUGGACUGUGGAAGUC-GCAUAAAUGUGCGCUUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAACCCGUCACAAGUAAU ((((........)))).((((-----(((((((((((.....))))-.)))..(((((.(((((((((...)))))))))...))))).............))))).)))... ( -29.50, z-score = -0.82, R) >consensus _______CGA_ACGGGAGCCCGUUG__AGGGAUCCUUGGGGAGGA__GCAUAAAUGUGCGCUUAAUUGAAUUAAUUAAGCCAACGCAUAAACAUAAAA_CCGUCACAAGUAAU .....................................................(((((.(((((((((...)))))))))...)))))......................... (-10.04 = -10.10 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:22 2011