
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,616,685 – 11,616,781 |
| Length | 96 |
| Max. P | 0.999006 |

| Location | 11,616,685 – 11,616,781 |
|---|---|
| Length | 96 |
| Sequences | 9 |
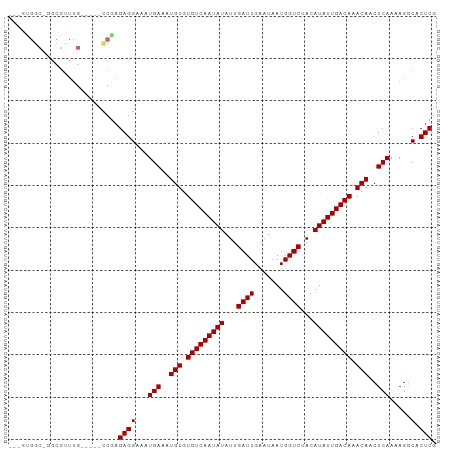
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 88.11 |
| Shannon entropy | 0.23398 |
| G+C content | 0.38502 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.66 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11616685 96 + 21146708 AUGGCGGCGGGCGUUUG-----UCGAGAGGAAAUGAAAUGUGUGUCAAUAUAUUGAUUGAAUAAUGGUCUACAUAUUGACAAACAACUCAAAAGGCACUCG .......((((.((((.-----((....))...(((..(((.(((((((((...((((.......))))...))))))))).)))..)))..)))).)))) ( -22.10, z-score = -1.09, R) >droSim1.chr2R 10352419 93 + 19596830 ---AUGGCGGGCGUUUG-----UCGAGAGGAAAUGAAAUGUGUGUCAAUAUAUUGAUUGAAUAAUGGUCUACAUAUUGACAAACAACUCAAAAGGCACUCG ---....((((.((((.-----((....))...(((..(((.(((((((((...((((.......))))...))))))))).)))..)))..)))).)))) ( -22.10, z-score = -1.66, R) >droSec1.super_1 9109955 93 + 14215200 ---AUGGCGGGCGUUUG-----UCGAGAGGAAAUGAAAUGUGUGUCAAUAUAUUGAUUGAAUAAUGGUCUACAUAUUGACAAACAACUCAAAAGGCACUCG ---....((((.((((.-----((....))...(((..(((.(((((((((...((((.......))))...))))))))).)))..)))..)))).)))) ( -22.10, z-score = -1.66, R) >droYak2.chr2R 3767657 98 - 21139217 ---GUGGCAGGCGUUUGGAAAGCCGAGAGGAAAUGAAAUGUGUGUCAAUAUAUUGAUUGAAUAAUGGUCUACAUAUUGACAAACAACUCAAAAGACACUCG ---......(((.........)))(((.(....(((..(((.(((((((((...((((.......))))...))))))))).)))..))).....).))). ( -22.80, z-score = -1.70, R) >droEre2.scaffold_4845 8343206 93 - 22589142 ---AUGGCAGGCGUUUG-----CCGAGAGGAAAUGAAAUGUGUGUCAAUAUAUUGAUUGAAUAAUGGUCUACAUAUUGACAAACAACUCAAAAGGCACUCG ---.((((((....)))-----))).((((...(((..(((.(((((((((...((((.......))))...))))))))).)))..))).....).))). ( -23.60, z-score = -2.08, R) >droAna3.scaffold_13266 7303241 82 - 19884421 -------------------AGGCCGAGAGGAAAUGAAAUGUGUGUCAAUAUAUUGAUUGAAUAAUGGUCUACAUAUUGACAAACAACUCAAAAGGCACUCG -------------------..(((..(((.........(((.(((((((((...((((.......))))...))))))))).))).)))....)))..... ( -18.80, z-score = -2.15, R) >dp4.chr3 207488 92 - 19779522 --GGUGGC--GCGUUUG-----CCAAGAGGAAAUGAAAUGUGUGUCAAUAUAUUGAUUGAAUAAUGGUCUACAUAUUGACAAACAACUCAAAAGGCACUCG --..((((--(....))-----))).((((...(((..(((.(((((((((...((((.......))))...))))))))).)))..))).....).))). ( -23.10, z-score = -1.93, R) >droPer1.super_37 186709 92 - 760358 --GGUGGC--GCGUUUG-----CCAAGAGGAAAUGAAAUGUGUGUCAAUAUAUUGAUUGAAUAAUGGUCUACAUAUUGACAAACAACUCAAAAGGCACUCG --..((((--(....))-----))).((((...(((..(((.(((((((((...((((.......))))...))))))))).)))..))).....).))). ( -23.10, z-score = -1.93, R) >droWil1.scaffold_180699 1599169 83 - 2593675 ------------------GGAAGGGUGAGGAAAUGAAAUGUGUGUCAAUAUAUUGAUUGAAUAAUGGUCUACAUAUUGACAAACAACUCAAAAGGCACUCG ------------------....(((((......(((..(((.(((((((((...((((.......))))...))))))))).)))..))).....))))). ( -21.90, z-score = -3.46, R) >consensus ___GUGGC_GGCGUUUG_____CCGAGAGGAAAUGAAAUGUGUGUCAAUAUAUUGAUUGAAUAAUGGUCUACAUAUUGACAAACAACUCAAAAGGCACUCG ..........................((((...(((..(((.(((((((((...((((.......))))...))))))))).)))..))).....).))). (-17.66 = -17.66 + -0.00)


| Location | 11,616,685 – 11,616,781 |
|---|---|
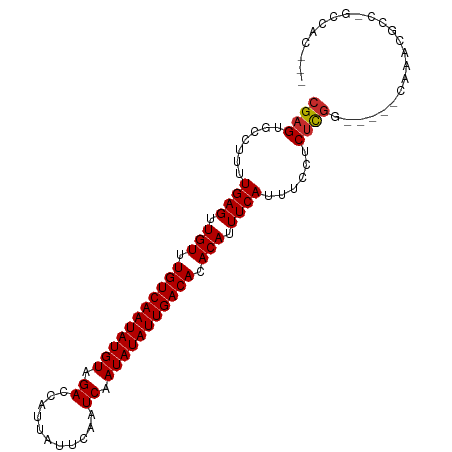
| Length | 96 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 88.11 |
| Shannon entropy | 0.23398 |
| G+C content | 0.38502 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -20.47 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11616685 96 - 21146708 CGAGUGCCUUUUGAGUUGUUUGUCAAUAUGUAGACCAUUAUUCAAUCAAUAUAUUGACACACAUUUCAUUUCCUCUCGA-----CAAACGCCCGCCGCCAU ((((.......((((.(((.(((((((((((.((...........)).))))))))))).))).))))......)))).-----................. ( -21.82, z-score = -3.47, R) >droSim1.chr2R 10352419 93 - 19596830 CGAGUGCCUUUUGAGUUGUUUGUCAAUAUGUAGACCAUUAUUCAAUCAAUAUAUUGACACACAUUUCAUUUCCUCUCGA-----CAAACGCCCGCCAU--- ((((.......((((.(((.(((((((((((.((...........)).))))))))))).))).))))......)))).-----..............--- ( -21.82, z-score = -3.88, R) >droSec1.super_1 9109955 93 - 14215200 CGAGUGCCUUUUGAGUUGUUUGUCAAUAUGUAGACCAUUAUUCAAUCAAUAUAUUGACACACAUUUCAUUUCCUCUCGA-----CAAACGCCCGCCAU--- ((((.......((((.(((.(((((((((((.((...........)).))))))))))).))).))))......)))).-----..............--- ( -21.82, z-score = -3.88, R) >droYak2.chr2R 3767657 98 + 21139217 CGAGUGUCUUUUGAGUUGUUUGUCAAUAUGUAGACCAUUAUUCAAUCAAUAUAUUGACACACAUUUCAUUUCCUCUCGGCUUUCCAAACGCCUGCCAC--- .(((.......((((.(((.(((((((((((.((...........)).))))))))))).))).))))......)))(((.............)))..--- ( -24.24, z-score = -3.92, R) >droEre2.scaffold_4845 8343206 93 + 22589142 CGAGUGCCUUUUGAGUUGUUUGUCAAUAUGUAGACCAUUAUUCAAUCAAUAUAUUGACACACAUUUCAUUUCCUCUCGG-----CAAACGCCUGCCAU--- .(((.......((((.(((.(((((((((((.((...........)).))))))))))).))).))))......)))((-----((......))))..--- ( -26.22, z-score = -4.60, R) >droAna3.scaffold_13266 7303241 82 + 19884421 CGAGUGCCUUUUGAGUUGUUUGUCAAUAUGUAGACCAUUAUUCAAUCAAUAUAUUGACACACAUUUCAUUUCCUCUCGGCCU------------------- ((((.......((((.(((.(((((((((((.((...........)).))))))))))).))).))))......))))....------------------- ( -22.02, z-score = -4.26, R) >dp4.chr3 207488 92 + 19779522 CGAGUGCCUUUUGAGUUGUUUGUCAAUAUGUAGACCAUUAUUCAAUCAAUAUAUUGACACACAUUUCAUUUCCUCUUGG-----CAAACGC--GCCACC-- .(((.(.....((((.(((.(((((((((((.((...........)).))))))))))).))).))))...)))).(((-----(......--))))..-- ( -23.70, z-score = -3.67, R) >droPer1.super_37 186709 92 + 760358 CGAGUGCCUUUUGAGUUGUUUGUCAAUAUGUAGACCAUUAUUCAAUCAAUAUAUUGACACACAUUUCAUUUCCUCUUGG-----CAAACGC--GCCACC-- .(((.(.....((((.(((.(((((((((((.((...........)).))))))))))).))).))))...)))).(((-----(......--))))..-- ( -23.70, z-score = -3.67, R) >droWil1.scaffold_180699 1599169 83 + 2593675 CGAGUGCCUUUUGAGUUGUUUGUCAAUAUGUAGACCAUUAUUCAAUCAAUAUAUUGACACACAUUUCAUUUCCUCACCCUUCC------------------ .(((.(.....((((.(((.(((((((((((.((...........)).))))))))))).))).))))...))))........------------------ ( -19.10, z-score = -4.18, R) >consensus CGAGUGCCUUUUGAGUUGUUUGUCAAUAUGUAGACCAUUAUUCAAUCAAUAUAUUGACACACAUUUCAUUUCCUCUCGG_____CAAACGCC_GCCAC___ ((((.......((((.(((.(((((((((((.((...........)).))))))))))).))).))))......))))....................... (-20.47 = -20.52 + 0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:20 2011