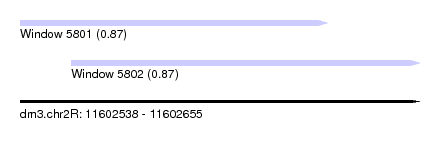
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,602,538 – 11,602,655 |
| Length | 117 |
| Max. P | 0.872518 |

| Location | 11,602,538 – 11,602,628 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.60 |
| Shannon entropy | 0.53763 |
| G+C content | 0.47492 |
| Mean single sequence MFE | -16.60 |
| Consensus MFE | -12.10 |
| Energy contribution | -11.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
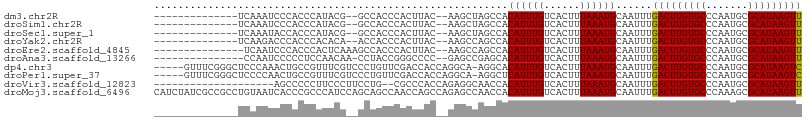
>dm3.chr2R 11602538 90 + 21146708 --------------UCAAAUCCCACCCAUACG--GCCACCCACUUAC--AAGCUAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUU --------------((((((...........(--((...........--......))).((((((......))))))..))))))(((((((.......))))))).. ( -14.93, z-score = -1.20, R) >droSim1.chr2R 10337249 90 + 19596830 --------------UCAAAUCCCACCCAUACG--GCCACCCACUUAC--AAGCUAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUU --------------((((((...........(--((...........--......))).((((((......))))))..))))))(((((((.......))))))).. ( -14.93, z-score = -1.20, R) >droSec1.super_1 9095878 90 + 14215200 --------------UCAAAUACCACCCAUACG--GCCACCCACUUAC--AAGCUAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUU --------------((((((...........(--((...........--......))).((((((......))))))..))))))(((((((.......))))))).. ( -15.53, z-score = -1.28, R) >droYak2.chr2R 3752717 90 - 21139217 --------------UCAAGACCCACCCACACA--ACCACCCACUUAC--AAGCCAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUU --------------..................--.............--..........((((((......))))))......(((((((((.......))))))))) ( -11.70, z-score = -0.88, R) >droEre2.scaffold_4845 8327507 91 - 22589142 ---------------UCAAUCCCACCCACUCAAAGCCACCCACUUAC--AAGCCAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUU ---------------................................--..........((((((......))))))......(((((((((.......))))))))) ( -11.70, z-score = -0.67, R) >droAna3.scaffold_13266 7290511 90 - 19884421 ---------------CCAAUCCCCCUCCAACAA-CCUACCGGGCCCC--GAGCCGAGCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUU ---------------..................-......((((..(--(((.((((..((((((......))))))...)))).)))).)))).............. ( -16.20, z-score = -0.68, R) >dp4.chr3 191769 102 - 19779522 -----GUUUCGGGCUCCCAAACUGCCGUUUCGUCCCUGUUCGACCACCAGGCA-AGGCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUC -----((((.((....))))))((((.....(((.......))).....))))-.((((((...((((..((......))..)))).))))))............... ( -24.70, z-score = -0.56, R) >droPer1.super_37 171142 102 - 760358 -----GUUUCGGGCUCCCCAACUGCCGUUUCGUCCCUGUUCGACCACCAGGCA-AGGCUCAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUC -----.....(((...)))...((((.....(((.......))).....))))-.(((......)))................(((((((((.......))))))))) ( -23.30, z-score = -0.29, R) >droVir3.scaffold_12823 1506195 86 + 2474545 --------------------AGCCCCCUUCCCUUCCUG--CGCCCACCAGAGGCAACCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUU --------------------................((--(((((....).)))......(((((......))))))))....(((((((((.......))))))))) ( -16.30, z-score = -1.14, R) >droMoj3.scaffold_6496 26459096 108 - 26866924 CAUCUAUCGCCGCCUGUAAUCACCCGCCCAUCCAGCAGCCAACCAGCCAGAGCCAACCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAAGCGCAUAAGUU ...........(((((.........((.......)).((......))))).))......((((((......))))))......(((((((((.......))))))))) ( -16.70, z-score = -0.25, R) >consensus ______________UCAAAUCCCACCCAUACG__GCCACCCACCUAC__AAGCCAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUU ...........................................................((((((......))))))......(((((((((.......))))))))) (-12.10 = -11.94 + -0.16)
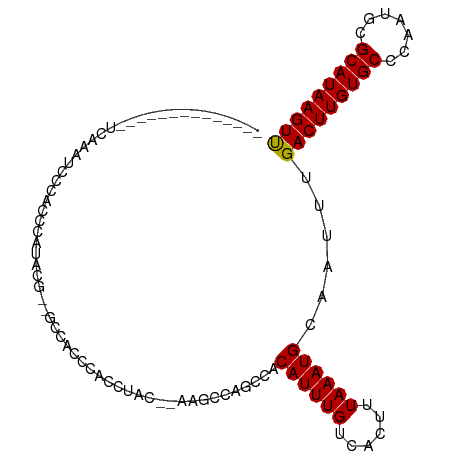

| Location | 11,602,553 – 11,602,655 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.01 |
| Shannon entropy | 0.54610 |
| G+C content | 0.46075 |
| Mean single sequence MFE | -17.87 |
| Consensus MFE | -11.89 |
| Energy contribution | -11.83 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
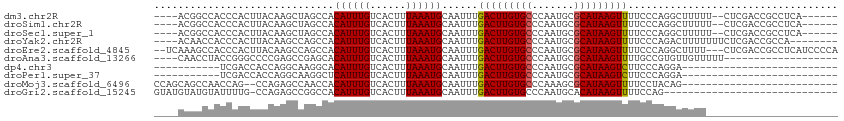
>dm3.chr2R 11602553 102 + 21146708 ----ACGGCCACCCACUUACAAGCUAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUUUUCCCAGGCUUUUU--CUCGACCGCCUCA------ ----..(((......((....))..((((.((((((......))))))......(((((((((.......)))))))))......))))....--.......)))...------ ( -20.10, z-score = -1.22, R) >droSim1.chr2R 10337264 102 + 19596830 ----ACGGCCACCCACUUACAAGCUAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUUUUCCCAGGCUUUUU--CUCGACCGCCUCA------ ----..(((......((....))..((((.((((((......))))))......(((((((((.......)))))))))......))))....--.......)))...------ ( -20.10, z-score = -1.22, R) >droSec1.super_1 9095893 102 + 14215200 ----ACGGCCACCCACUUACAAGCUAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUUUUCCCAGGCUUUUU--CUCGACCGCCUCA------ ----..(((......((....))..((((.((((((......))))))......(((((((((.......)))))))))......))))....--.......)))...------ ( -20.10, z-score = -1.22, R) >droYak2.chr2R 3752732 102 - 21139217 ----ACAACCACCCACUUACAAGCCAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUUUUCCCAGACUUUUUUUCUCGACCGCCA-------- ----......................((..((((((......))))))......(((((((((.......))))))))).......................))..-------- ( -12.70, z-score = -0.62, R) >droEre2.scaffold_4845 8327521 109 - 22589142 --UCAAAGCCACCCACUUACAAGCCAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUUUUCCCAGGCUUUU---CUCGACCGCCUCAUCCCCA --..((((((....................((((((......))))))......(((((((((.......)))))))))......)))))).---................... ( -19.60, z-score = -1.95, R) >droAna3.scaffold_13266 7290526 91 - 19884421 ----CAACCUACCGGGCCCCGAGCCGAGCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUUUUGCCGUGUUGUUUUU------------------- ----..........(((.....))).((((((((((......)))))((((...(((((((((.......))))))))))))).)))))......------------------- ( -21.70, z-score = -0.90, R) >dp4.chr3 191803 77 - 19779522 -----------UCGACCACCAGGCAAGGCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUCUUCCCAGGA-------------------------- -----------.......((.((((((.....))))))................(((((((((.......)))))))))......)).-------------------------- ( -18.70, z-score = -1.10, R) >droPer1.super_37 171176 77 - 760358 -----------UCGACCACCAGGCAAGGCUCAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUCUUCCCAGGA-------------------------- -----------.......((.((((((.....))))))................(((((((((.......)))))))))......)).-------------------------- ( -18.70, z-score = -1.09, R) >droMoj3.scaffold_6496 26459127 86 - 26866924 CCAGCAGCCAACCAG--CCAGAGCCAACCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAAGCGCAUAAGUUUUCCUACAG-------------------------- ...(((.........--.((((..........))))..........))).....(((((((((.......))))))))).........-------------------------- ( -12.35, z-score = -0.13, R) >droGri2.scaffold_15245 12561618 84 - 18325388 GUAUGUAUGUAUUUUG-CCAGAGCCGGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCACAUAAGUUUUCCAG----------------------------- .(((((..(((((..(-(....)).((.((((...((((..((......))..)))).)))).))))))))))))..........----------------------------- ( -14.70, z-score = 0.20, R) >consensus ____ACAGCCACCCACUUACAAGCCAGCCACAUUUGUCACUUUAAAUGCAAUUUGACUUGUGCCCAAUGCGCAUAAGUUUUCCCAGGCUUUU______________________ ..............................((((((......))))))......(((((((((.......)))))))))................................... (-11.89 = -11.83 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:16 2011