
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,602,272 – 11,602,371 |
| Length | 99 |
| Max. P | 0.874923 |

| Location | 11,602,272 – 11,602,371 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.13 |
| Shannon entropy | 0.58903 |
| G+C content | 0.57781 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -13.21 |
| Energy contribution | -13.56 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11602272 99 + 21146708 --------CAUUCCCCGCCCCUUUGGGCCCACAAGUGCUGACAGCGAUAAAUUCUAAUGCCG-GGUCAGCACUGCCAGC-GACCACGCCUCCG-AUUCCAGGGCGUAUGU-- --------.......(((((.....(((.....(((((((((.(((.((.....)).)))..-.)))))))))))).((-(....))).....-......))))).....-- ( -32.20, z-score = -1.15, R) >droSim1.chr2R 10336983 99 + 19596830 --------CAUACCCCGCCCCUUUGGGCCCACAAGUGCUGACAGCGAUAAAUUCUAAUGCCG-GGUCAGCACUGGCAGC-GACCACGCCUCCG-AUUCCAGGGCGUAUGU-- --------((((((((((((....)))).....(((((((((.(((.((.....)).)))..-.)))))))))((..((-(....)))..)).-......))).))))).-- ( -38.60, z-score = -2.64, R) >droSec1.super_1 9095612 99 + 14215200 --------CAUUCCCCGCCCCUUUGGGCCCACAAGUGCUGACAGCGAUAAAUUCUAAUGCCG-GGUCAGCACUGGCAGC-GACCACGCCUCCG-AUUCCAGGGCGUAUGU-- --------.......(((((..(((((((....(((((((((.(((.((.....)).)))..-.)))))))))))).((-(....)))..)))-).....))))).....-- ( -35.30, z-score = -1.59, R) >droYak2.chr2R 3752448 101 - 21139217 --------CUUUCCCCGCCCCUUUGGACCCACAAGUGCUGACAGCGAUAAAUUCUAAUGCCG-GGUCAGCACUGCCAGC-GACCACGCCUUCG-AUUCCAGGGCGCGUAUGU --------.......(((((((..(((..(...(((((((((.(((.((.....)).)))..-.)))))))))....((-(....)))....)-..))))))).)))..... ( -32.30, z-score = -1.56, R) >droEre2.scaffold_4845 8327241 99 - 22589142 --------CUUUCCCCGCCCCUUUGGCCCCACAAGUGCUGACAGCGAUAAAUUCUAAUGCCG-GGUCAGCACAGCCAGC-GACCACGCCUCCG-UUUCCAGGGCGUAUGU-- --------.......(((((...((((.......((((((((.(((.((.....)).)))..-.)))))))).))))((-(....))).....-......))))).....-- ( -30.20, z-score = -1.23, R) >dp4.chr3 191415 107 - 19779522 AGGCACCACAAGAAGACCUUCCUCCUUUUCCCAAGUGCUGACAGCGAUAAAUUCUAAUGCCG-GGUCAGCACUGCCAGC-GACCACGCCUCCC-UCUGAGGCACCCGCUC-- .(((.....(((.((......)).)))......(((((((((.(((.((.....)).)))..-.))))))))))))(((-(.....(((((..-...)))))...)))).-- ( -33.40, z-score = -2.50, R) >droPer1.super_37 170789 107 - 760358 AGGCACCACAAGAAGACCUUCCUCCUUUUCCCAAGUGCUGACAGCGAUAAAUUCUAAUGCCG-GGUCAGCACUGCCAGC-GACCACGCCUCCC-UCUGAGGCACCCGCUC-- .(((.....(((.((......)).)))......(((((((((.(((.((.....)).)))..-.))))))))))))(((-(.....(((((..-...)))))...)))).-- ( -33.40, z-score = -2.50, R) >droWil1.scaffold_180699 1574347 91 - 2593675 -------------------AAAACUUCGUUCCAUGUGCUGACAGCGAUAAAUUCUAAUGCCGAGGUCAGCACUACCCCUUGACCACGCCUCCCAUCCCCCUUACAUGUGU-- -------------------......(((......((((((((..((..............))..)))))))).......)))............................-- ( -15.66, z-score = -1.13, R) >droGri2.scaffold_15245 12561298 81 - 18325388 ----------------AACAUAGCGCUCCUCCAUGUGCUGACAGCGAUAAAUUCUAAUGUCG-GGUCA----UGUUGGU-GGCCACGCCUCCG-GC----GGGUGUGG---- ----------------........(((((..((((..((((((..((.....))...)))))-)..))----))..)).-)))(((((((...-..----))))))).---- ( -27.60, z-score = -0.66, R) >consensus ________CAUUCCCCGCCCCUUUGGGCCCACAAGUGCUGACAGCGAUAAAUUCUAAUGCCG_GGUCAGCACUGCCAGC_GACCACGCCUCCG_AUUCCAGGGCGUGUGU__ ..................................((((((((.(((.((.....)).)))....))))))))........................................ (-13.21 = -13.56 + 0.35)

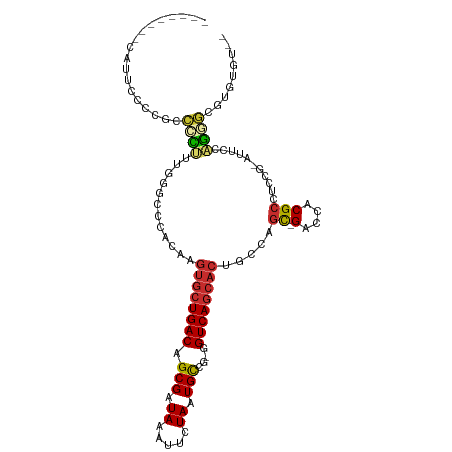
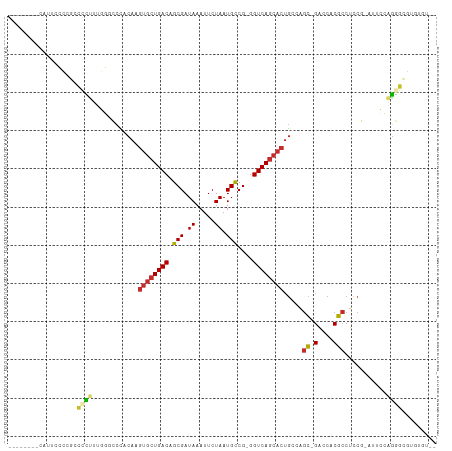
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:15 2011