
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,553,955 – 11,554,021 |
| Length | 66 |
| Max. P | 0.831861 |

| Location | 11,553,955 – 11,554,021 |
|---|---|
| Length | 66 |
| Sequences | 8 |
| Columns | 71 |
| Reading direction | forward |
| Mean pairwise identity | 65.07 |
| Shannon entropy | 0.68377 |
| G+C content | 0.53601 |
| Mean single sequence MFE | -18.21 |
| Consensus MFE | -4.69 |
| Energy contribution | -5.09 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
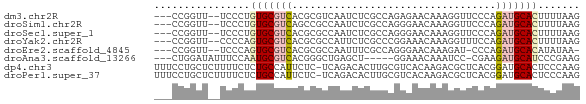
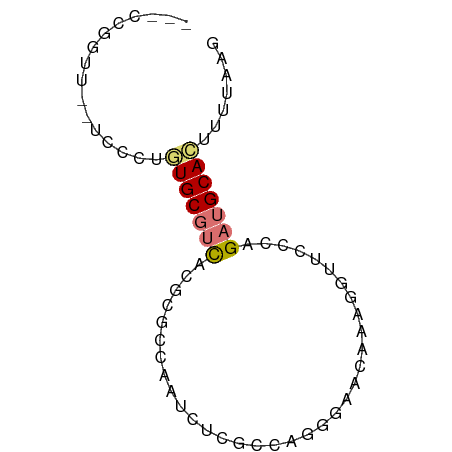
>dm3.chr2R 11553955 66 + 21146708 ---CCGGUU--UCCCUGUGCGUCACGCGUCAAUCUCGCCAGAGAACAAAGGUUCCCAGAUGCACUUUUAAG ---..((..--..)).(((((((..(((.......)))..(.((((....)))).).)))))))....... ( -17.00, z-score = -1.73, R) >droSim1.chr2R 10285651 66 + 19596830 ---CCGGUU--UCCCUGUGCGUCAGCCGCCAAUCUCGCCAGGGAACAAAGGUUCCCAGAUGCACUUUUAAG ---..((..--..)).(((((((....((.......))..((((((....)))))).)))))))....... ( -20.90, z-score = -2.21, R) >droSec1.super_1 9047045 66 + 14215200 ---CCGGUU--UCCCUGUGCGUCACGCGCCAAUCUCGCCAGGGAACAAAGGUUCCCAGAUGCACUUUUAAG ---..((..--..)).(((((((..(((.......)))..((((((....)))))).)))))))....... ( -22.90, z-score = -2.88, R) >droYak2.chr2R 3702582 66 - 21139217 ---CCGGUU--CCCCAGUGCGUCACGCGCCAUUCUCGCCCGGAAACAAAGGUUUCCAGAUGCACUUUUAAG ---..((..--..))((((((((..(((.......)))..((((((....)))))).))))))))...... ( -21.90, z-score = -3.42, R) >droEre2.scaffold_4845 8279468 64 - 22589142 ---CCGGUU--UCCCAGUGCGUCACGCGCCAAUUUCGCCAGGGAACAAAGAU-CCCAGAUGCACAUAUAA- ---..((..--..)).(((((((..(((.......)))..((((.......)-))).)))))))......- ( -19.60, z-score = -2.84, R) >droAna3.scaffold_13266 7251932 62 - 19884421 ---CUGGAUAUUUCCAAUGCGUCACGGGCUGAGCU-----GGAAACAAAUCC-CGAAGAUGCAUCCCGAAG ---.((((....))))(((((((.((((......(-----(....))...))-))..)))))))....... ( -19.40, z-score = -2.23, R) >dp4.chr3 148566 70 - 19779522 UUUCCUGCUCUUUUCUCUGCCAUUCUC-UCAGACACUUGCGUCACAAGACGCUCACGGAUGCACUCCCAAG .....((((((....((((........-.)))).....(((((....)))))....))).)))........ ( -12.00, z-score = -1.12, R) >droPer1.super_37 128065 70 - 760358 UUUCCUGCUCUUUUCUCUGCCAUUCUC-UCAGACACUUGCGUCACAAGACGCUCACGGAUGCACUCCCAAG .....((((((....((((........-.)))).....(((((....)))))....))).)))........ ( -12.00, z-score = -1.12, R) >consensus ___CCGGUU__UCCCUGUGCGUCACGCGCCAAUCUCGCCAGGGAACAAAGGUUCCCAGAUGCACUUUUAAG ................(((((((..................................)))))))....... ( -4.69 = -5.09 + 0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:11 2011