| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,550,367 – 11,550,459 |
| Length | 92 |
| Max. P | 0.615503 |

| Location | 11,550,367 – 11,550,459 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 73.45 |
| Shannon entropy | 0.48356 |
| G+C content | 0.65859 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -21.75 |
| Energy contribution | -22.76 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.615503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11550367 92 + 21146708 GGCGACUGAGCGGUACCGGCGGCUACGGUCGUCGGCGACUGACCCGCCGAACUGAACAUACCCGUAGUGGC---GGCCAGCAGUGAUUGCUGCUU--- (((.......((((.((((((((....))))))))..))))...(((((...((........))...))))---))))((((((....)))))).--- ( -39.20, z-score = -1.18, R) >droWil1.scaffold_181009 2038881 71 - 3585778 GCCGACUGGG------CGCCGGCUGGUGGCGGCGGCGGCUGAGCU---------AACAUACCCGGAGAG---------AGCAGCGACGGCUGCUU--- .....(((((------(((((.(....).)))))((......)).---------......)))))...(---------((((((....)))))))--- ( -31.10, z-score = -0.26, R) >dp4.chr3 144991 71 - 19779522 GCCGACUGGG------CGCCGGUUGGAGUCGGCGGCGACUGAGCU---------AACAUACCCGUGGAU---------AGCAGCGAGGGCUGCUU--- .((((((((.------..)))))))).(((.((((.(..((....---------..))..))))).)))---------((((((....)))))).--- ( -27.90, z-score = -0.39, R) >droAna3.scaffold_13266 7248515 98 - 19884421 GGCGACUGGGAGGCAGUGCCAGUAGAGGCCGAUGCCGAUUGACCUGCCGAACUGAACAUACCCGAGGUGGCCGCAGCCAGCAGCGAGGGCUGCUUAUG (((.((((((.(((((.(.((((...(((....))).)))).))))))(.......)...)))..))).)))((((((.........))))))..... ( -38.70, z-score = -1.18, R) >droEre2.scaffold_4845 8275859 92 - 22589142 GGCGACUGAGCGGUACCGGCGGCUACGGUCGUCGGCGACUGACCCGCCGAACUGAACAUACCCGUAGUGGC---GGCCAGCAGUGAUUGCUGCUU--- (((.......((((.((((((((....))))))))..))))...(((((...((........))...))))---))))((((((....)))))).--- ( -39.20, z-score = -1.18, R) >droYak2.chr2R 3699017 92 - 21139217 GGCGACUGAGCGGUACCGGCGGCUACGGUCGUCGGCGACUGACCCGCCGAACUAAACAUACCCGUAGUGGC---GGCCAGCAGUGAUUGCUGCUU--- (((.......((((.((((((((....))))))))..))))...((((..((((..........)))))))---))))((((((....)))))).--- ( -38.90, z-score = -1.27, R) >droSec1.super_1 9043448 92 + 14215200 GGCGACUGAGCGGUACCGGCGACUACGGUCGUCGGCGACUGACCCGCCGAACUGAACAUACCCGUAGUGGC---GGCCAGCAGUGAUUGCUGCUU--- (((.......((((.((((((((....))))))))..))))...(((((...((........))...))))---))))((((((....)))))).--- ( -39.80, z-score = -2.02, R) >consensus GGCGACUGAGCGGUACCGGCGGCUACGGUCGUCGGCGACUGACCCGCCGAACUGAACAUACCCGUAGUGGC___GGCCAGCAGUGAUUGCUGCUU___ ((((..(.((.....((((((((....))))))))...)).)..))))..............................((((((....)))))).... (-21.75 = -22.76 + 1.01)

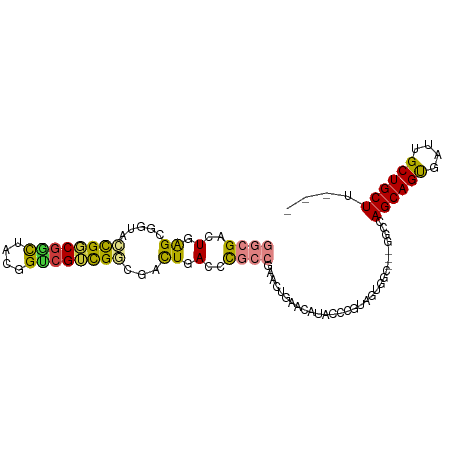
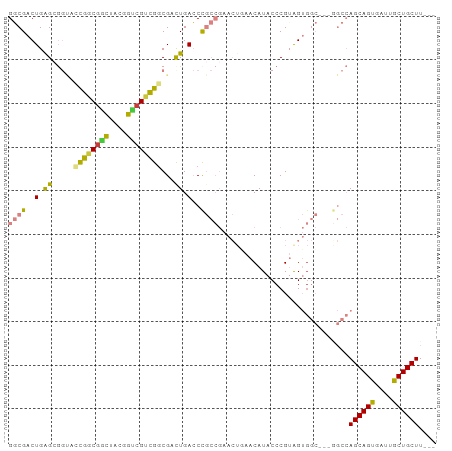
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:10 2011