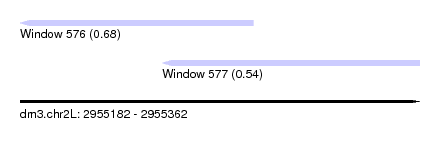
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,955,182 – 2,955,362 |
| Length | 180 |
| Max. P | 0.678163 |

| Location | 2,955,182 – 2,955,287 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.39 |
| Shannon entropy | 0.18921 |
| G+C content | 0.50714 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -33.93 |
| Energy contribution | -33.97 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.678163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2955182 105 - 23011544 GACGUGAGCCGCCAGAGUUCCAGCUUUGGUUGUCAUAA---------------AAGCCCCUAGAUUGGUUGGGAUAAAGCCAAGAUAGCGGUGGUUGUGGUAGAUCCCCUUUAUUGGGCU ......((((((((((((....))))))))(..(((((---------------..(((.(((..((((((.......))))))..))).)))..)))))..)..............)))) ( -36.00, z-score = -1.86, R) >droSec1.super_5 1111195 120 - 5866729 GAUGUGAGCCGCCAGAGUUCCAGCUUUGGUUAUCACAAAAGUCUGGAUUUCGGAAGCCCCCAGAUUGGCUGGAAUAAAGCCAAGAUUGCGGUGGUUGUGGUAGAUCCUCUUUCUUGGGCU ......((((((((((((....))))))))...((..((((...(((((((.(.(((((((((.((((((.......))))))..))).)).)))).).).)))))).))))..)))))) ( -42.00, z-score = -1.57, R) >droSim1.chr2L 2913564 120 - 22036055 GAUGUGAGCCGCCAGAGUUCCAGCUUUGGUUAUCACAAAAGUCUGGAUUCCGGAAGCCCCCAGAUUGGCUGGGAUAAAGCCAAGAUUGCGGUGGUUGUGGUAGAUCCUCUUUCUUGGGCU ......((((((((((((....))))))))...((..((((...((((((((..(((((((((.((((((.......))))))..))).)).)))).)))..))))).))))..)))))) ( -42.60, z-score = -1.25, R) >consensus GAUGUGAGCCGCCAGAGUUCCAGCUUUGGUUAUCACAAAAGUCUGGAUU_CGGAAGCCCCCAGAUUGGCUGGGAUAAAGCCAAGAUUGCGGUGGUUGUGGUAGAUCCUCUUUCUUGGGCU ......((((((((((((....))))))))((((((.....((((.....))))(((((((((.((((((.......))))))..))).)).))))))))))..............)))) (-33.93 = -33.97 + 0.04)



| Location | 2,955,246 – 2,955,362 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.58 |
| Shannon entropy | 0.26519 |
| G+C content | 0.37800 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -23.30 |
| Energy contribution | -22.14 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2955246 116 - 23011544 AAAUUCGGUUAAUUUAAUUAUUUAAAUGAAUAAUUGAUUUUCAGCUUACUG---GUGCUUCCCACUGCGACUGAUUCUGACGUGAGCCGCCAGAGUUCCAGCUUUGGUUGUCAUAAAAG- ....((((((...((((((((((....))))))))))......((....((---(......)))..)))))))).......((((.(.((((((((....)))))))).))))).....- ( -27.00, z-score = -1.58, R) >droEre2.scaffold_4929 3000771 117 - 26641161 AAAUUGGAUUAAUUUAAUUGUUUAAAUGAUUAGUUGGUUGUCAGCUUACUG---GUGCUUUCCACUGCGGUUGAUUUUGAUAUGGGACGCCAGAGUUCCAGCUUUGGUUACCAUAAAAGU ......((((((((.(((..(....)..)))))))))))(((((((((.((---(......))).)).))))))).....(((((...((((((((....))))))))..)))))..... ( -28.70, z-score = -1.10, R) >droYak2.chr2L 2948522 117 - 22324452 AAAUUUGUUUAUUUAAAUUGUUCAAAUGGUUAGUGGGGUUUCGGCUUACU---UGUGCUGUGCACUGCGGUUGAUUUUGAUAUGGGACGCCAGAAUUCCAGCUUUGGUUACCAUAAACGU .((((((......))))))..(((((......(..(.((..((((.....---...)))).)).)..).......)))))(((((...((((((........))))))..)))))..... ( -24.42, z-score = 0.82, R) >droSec1.super_5 1111273 120 - 5866729 AAAUUCCGUUAAUUUAAUUAUUUAAAUGAAUAAUUGGUUUUCAGCUUACUGCUUGUGUUUGCCACUGCGGCUGAUUCUGAUGUGAGCCGCCAGAGUUCCAGCUUUGGUUAUCACAAAAGU .............((((((((((....))))))))))...((((((....((..(((.....))).))))))))..((..(((((...((((((((....))))))))..)))))..)). ( -31.10, z-score = -2.09, R) >droSim1.chr2L 2913642 120 - 22036055 AAAUUCCGUUAAUUUAAUUAUUUAAAUGAAUAAUUGGUUUUCAGCUUACUGCUUGUGUUUGCCACUGCGGCUGAUUCUGAUGUGAGCCGCCAGAGUUCCAGCUUUGGUUAUCACAAAAGU .............((((((((((....))))))))))...((((((....((..(((.....))).))))))))..((..(((((...((((((((....))))))))..)))))..)). ( -31.10, z-score = -2.09, R) >consensus AAAUUCGGUUAAUUUAAUUAUUUAAAUGAAUAAUUGGUUUUCAGCUUACUG__UGUGCUUGCCACUGCGGCUGAUUCUGAUGUGAGCCGCCAGAGUUCCAGCUUUGGUUAUCAUAAAAGU .............((((((((((....))))))))))...((((((.((.....))((........))))))))......(((((...((((((((....))))))))..)))))..... (-23.30 = -22.14 + -1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:39 2011