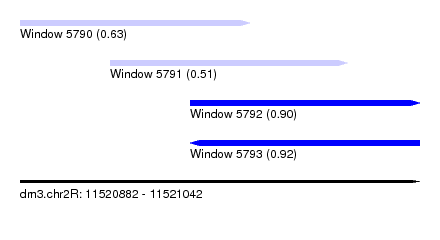
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,520,882 – 11,521,042 |
| Length | 160 |
| Max. P | 0.916889 |

| Location | 11,520,882 – 11,520,974 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 62.40 |
| Shannon entropy | 0.71381 |
| G+C content | 0.50917 |
| Mean single sequence MFE | -18.84 |
| Consensus MFE | -4.06 |
| Energy contribution | -4.56 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.632124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11520882 92 + 21146708 CUGCAGCAUAUUCACAACUUGCCAGAUACUUUUCUGGCUCAUCCAGCUGCCAUCCACUCACA--CUUGCCA------GCAUUCUGCACACACA------UCGUGAG .(((((.((...........((((((......)))))).......((((.((..........--..)).))------)))).)))))(((...------..))).. ( -23.30, z-score = -2.91, R) >droSim1.chr2R 10252243 84 + 19596830 CUGCAGCAUAUUCACAACUUGCCAGAUACUUUUCUGGCUCAUCCAGCUGCCAUC-AUCCACU--CCCACCAGC-------------ACACACA------UCGUGAG ...........((((.....((((((......)))))).......((((.....-.......--.....))))-------------.......------..)))). ( -17.93, z-score = -2.59, R) >droSec1.super_1 9014182 97 + 14215200 CUGCAGCAUAUUCACAACUUGCCAGAUACUUUUCUGGCUCAUCCAGCUUCCAUC-AUCCACU--CCCACCAGCGCCAGCGUUCUGCACACACA------UCGUGAG .(((((.((...........((((((......))))))................-.......--.......((....)))).)))))(((...------..))).. ( -19.90, z-score = -1.98, R) >droYak2.chr2R 3669219 105 - 21139217 CUGCAGCAUAUUCACAACUUGCCAGAUACUUUUCUGGCUCAUCCAGCUGCCAUCCACUCACAUCCCUGCCAUC-CUUGCGCUCUGCACACACACACACAUCGUGAG ..(((((.............((((((......)))))).......)))))......(((((......((....-...))((...))...............))))) ( -21.65, z-score = -1.97, R) >droEre2.scaffold_4845 8246713 94 - 22589142 CUGCAGCAUAUUCACAACUUGCCAGAUACUUUUCUGGCUCAUCCAGCUGCCAUCCACUCCCA--CUUGCCGGC------AUUCUGCACACACACA----UCGUGAG .(((((.((...........((((((......)))))).......((((.((..........--..)).))))------)).)))))..(((...----..))).. ( -22.60, z-score = -2.39, R) >droAna3.scaffold_13266 7209836 73 - 19884421 CUGCAGCUCAUUCACAACUUUCCAGAUACUUUUCUGGCU--UUCAGGUUUCUUCUUUUCCCUG-UUUUCGAGUGAG------------------------------ ......(((((((........(((((......)))))..--..((((............))))-.....)))))))------------------------------ ( -17.20, z-score = -2.67, R) >dp4.chr3 113733 91 - 19779522 CUGCAGCAUAUUCACAACUUUCCAGAUACGUUUCUUUCCUCAACCAGAGCCCCCGCCCCUACAACGCUCUUCUGAGCAGACACAUCGCUGG--------------- ...((((..........((....))....((.(((...((((...(((((...............)))))..)))).))).))...)))).--------------- ( -15.16, z-score = -1.36, R) >droPer1.super_37 92701 91 - 760358 CUGCAGCAUAUUCACAACUUUCCAGAUACGUUUCUUUCCCCAACCAGAGCCCCCGCCCCUACAACGCUCUUCUGAGCAGACACAUCGCUGG--------------- ...((((..........((....))....((.(((.............((....)).........((((....))))))).))...)))).--------------- ( -13.00, z-score = -0.73, R) >consensus CUGCAGCAUAUUCACAACUUGCCAGAUACUUUUCUGGCUCAUCCAGCUGCCAUCCACCCACA__CCCGCCAGC______ACUCUGCACACACA______UCGUGAG .....................(((((......)))))..................................................................... ( -4.06 = -4.56 + 0.50)
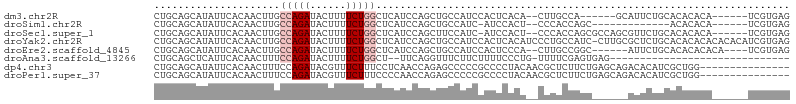

| Location | 11,520,918 – 11,521,013 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.55 |
| Shannon entropy | 0.57095 |
| G+C content | 0.46191 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -8.76 |
| Energy contribution | -9.29 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.506880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11520918 95 + 21146708 -GCUCAUCCAG--CUGCCAUCCACUCACA--CUUGCCA-----GCA------UUCUGCACACACAUCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAU -.........(--(((.((..........--..)).))-----)).------.(((((.(.((.(((....))))).)))))).((((((((((.......)))))))))) ( -26.70, z-score = -2.34, R) >droSim1.chr2R 10252279 87 + 19596830 -GCUCAUCCAG--CUGCCAUC-AUCCACU--CCCACCA-----GC-------------ACACACAUCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAU -.........(--((((.(((-.....((--......)-----).-------------.(((.....))).))).)))))....((((((((((.......)))))))))) ( -20.00, z-score = -1.53, R) >droSec1.super_1 9014218 100 + 14215200 -GCUCAUCCAG--CUUCCAUC-AUCCACU--CCCACCA-----GCGCCAGCGUUCUGCACACACAUCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAU -(((.....))--).......-.......--.......-----((....))..(((((.(.((.(((....))))).)))))).((((((((((.......)))))))))) ( -24.20, z-score = -1.93, R) >droYak2.chr2R 3669255 108 - 21139217 -GCUCAUCCAG--CUGCCAUCCACUCACAUCCCUGCCAUCCUUGCGCUCUGCACACACACACACAUCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAU -..........--(((((.....(((((......((.......))((...))...............))))).....)))))..((((((((((.......)))))))))) ( -24.20, z-score = -1.11, R) >droEre2.scaffold_4845 8246749 97 - 22589142 -GCUCAUCCAG--CUGCCAUCCACUCCCA--CUUGCCG-----GCAUU----CUGCACACACACAUCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAU -(((.....))--)...............--.((((((-----.((.(----((.(((.........)))))).))))))))..((((((((((.......)))))))))) ( -25.00, z-score = -1.50, R) >droAna3.scaffold_13266 7209872 76 - 19884421 ---GCUUUCAG--GUUUCUUCUUUUCCCUG-UUUUCGA-----------------------------GUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAU ---(((....(--((((((((.........-.....))-----------------------------).))))))..)))....((((((((((.......)))))))))) ( -14.54, z-score = -0.44, R) >dp4.chr3 113769 94 - 19779522 UCCUCAACCAGAGCCCCCGCCCCUACAACGCUCUUCUGA-----------------GCAGACACAUCGCUGGAUGUCGGUAGAUAUUUGCAUUUUAUGUCUAUAUGCAAAU ..(((.....))).........((((...((((....))-----------------))...((((((....))))).)))))..((((((((...........)))))))) ( -20.90, z-score = -1.29, R) >droPer1.super_37 92737 94 - 760358 UCCCCAACCAGAGCCCCCGCCCCUACAACGCUCUUCUGA-----------------GCAGACACAUCGCUGGAUGGCGGUAGAUAUUUGCAUUUUAUGUCUAUAUGCAAAU .(((((.((((.((....)).........((((....))-----------------))..........)))).))).)).....((((((((...........)))))))) ( -23.50, z-score = -1.64, R) >consensus _GCUCAUCCAG__CUGCCAUCCAUUCACA__CCUGCCA_____GC___________GCACACACAUCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAU .................................................................((((......)))).....((((((((((.......)))))))))) ( -8.76 = -9.29 + 0.53)

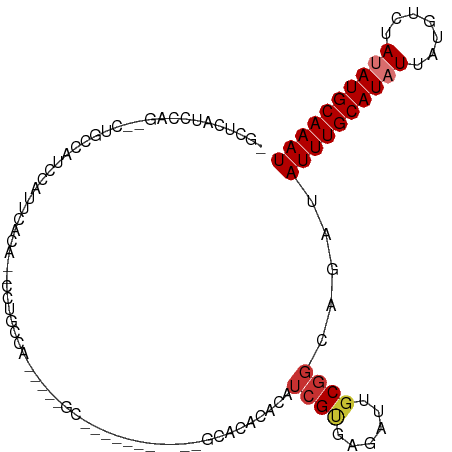
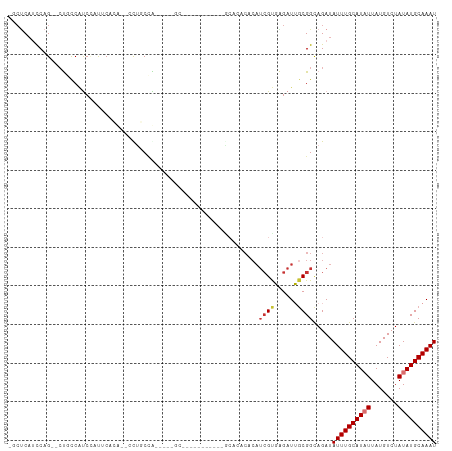
| Location | 11,520,950 – 11,521,042 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 64.43 |
| Shannon entropy | 0.65524 |
| G+C content | 0.45459 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -8.24 |
| Energy contribution | -8.47 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.902961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11520950 92 + 21146708 -----AGCAUUCUGCACACACA------UCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAUGAUGGCGC-GGAGCGCAGGAGCAAAGGAGC------------ -----.((.((((((.(.....------((....))(((((.((..(((((((((((.......))))))))))).)).)))-)).).))))))))........------------ ( -34.00, z-score = -3.31, R) >droSim1.chr2R 10252311 84 + 19596830 -------------GCACACACA------UCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAUGAUGGCGC-GGAGCGCAGGAGCAAAGGAGC------------ -------------((.......------((....))(((((.((..(((((((((((.......))))))))))).)).)))-)).))((....))........------------ ( -26.00, z-score = -1.81, R) >droSec1.super_1 9014255 92 + 14215200 -----AGCGUUCUGCACACACA------UCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAUGAUGGCGC-GGAGCGCAGGAGCAAAGGCGC------------ -----.(((((((((.(.((.(------((....))))).)((...(((((((((((.......)))))))))))...))))-)))))))....((....))..------------ ( -36.50, z-score = -3.20, R) >droYak2.chr2R 3669289 96 - 21139217 AUCCUUGCGCUCUGCACACACACACACAUCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAUGAUGGCCCAGGAGCAAAGGAGC-------------------- .(((((..((((((((..(.(((.......))).)..)))(((...(((((((((((.......)))))))))))...)))..))))).)))))..-------------------- ( -34.20, z-score = -3.22, R) >droEre2.scaffold_4845 8246781 94 - 22589142 -----GGCAUUCUGCACACACACA----UCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAUGAUGGCGC-GGAGCGCAGGAGCAAGGGAGA------------ -----.((.((((((.(.......----((....))(((((.((..(((((((((((.......))))))))))).)).)))-)).).))))))))........------------ ( -33.80, z-score = -3.22, R) >droAna3.scaffold_13266 7209896 69 - 19884421 ---------------GUUUUCG-------AGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAUGAUGGG---GGAGCGCUUGA---------------------- ---------------....(((-------((((...((.(..((..(((((((((((.......))))))))))).)).)---.)).)))))))---------------------- ( -19.20, z-score = -1.80, R) >dp4.chr3 113799 104 - 19779522 ------CUCUUCUGAGCAGACACAU-CGC---UGGAUGUCGGUAGAUAUUUGCAUUUUAUGUCUAUAUGCAAAU-CUC-UACAGGAGACCAAAAGAGGAGCAGGAAAAGUGGAGUA ------(((((((.(((.((....)-)))---)((.(.((.(((((.((((((((...........))))))))-.))-)))..)).)))...)))))))................ ( -27.10, z-score = -0.89, R) >droPer1.super_37 92767 104 - 760358 ------CUCUUCUGAGCAGACACAU-CGC---UGGAUGGCGGUAGAUAUUUGCAUUUUAUGUCUAUAUGCAAAU-CUC-UACAGGAGACCAAAAGAGGAGCAGGAAAAGUGGAGUA ------(((((((............-(((---(....))))(((((.((((((((...........))))))))-.))-))).(....)....)))))))................ ( -26.60, z-score = -0.72, R) >droGri2.scaffold_15112 3823763 87 - 5172618 --------------AACAUACACAU-GGCCUUUAGAUUGCGCCACAUAUUUGCAUAUGAUGUCUAUAUGCAAAU-AUAACAGCAGAAGCCAAAGCAGAAGCUG------------- --------------..........(-(((.(((....(((.....(((((((((((((.....)))))))))))-))....)))))))))).(((....))).------------- ( -22.50, z-score = -2.28, R) >consensus ______GC_UUCUGCACACACACA____UCGUGAGAUUGCGGCAGAUAUUUGCAUAUUAUGUCUAUAUGCAAAUGAUGGCGC_GGAGCGCAGGAGCAAAGGAG_____________ ...............................................((((((((((.......)))))))))).......................................... ( -8.24 = -8.47 + 0.22)

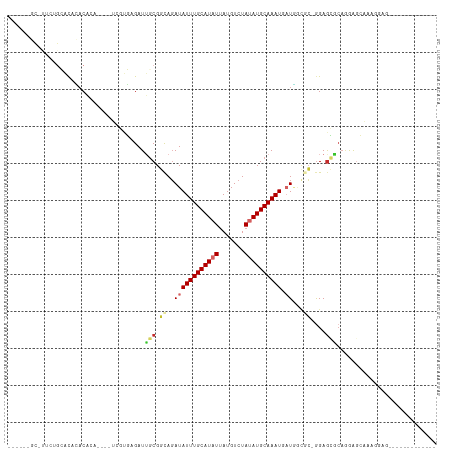
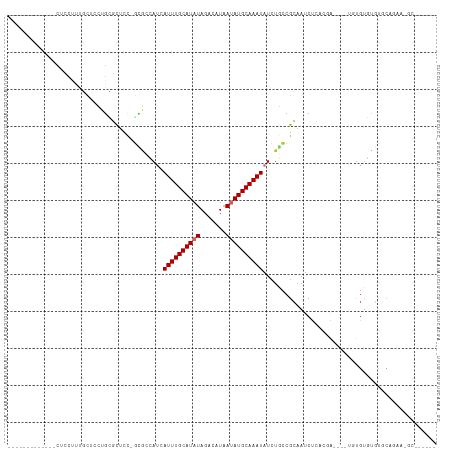
| Location | 11,520,950 – 11,521,042 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 64.43 |
| Shannon entropy | 0.65524 |
| G+C content | 0.45459 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -8.22 |
| Energy contribution | -8.44 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11520950 92 - 21146708 ------------GCUCCUUUGCUCCUGCGCUCC-GCGCCAUCAUUUGCAUAUAGACAUAAUAUGCAAAUAUCUGCCGCAAUCUCACGA------UGUGUGUGCAGAAUGCU----- ------------((......))....((((...-))))....((((((((((.......)))))))))).((((((((((((....))------).)))).))))).....----- ( -27.10, z-score = -2.28, R) >droSim1.chr2R 10252311 84 - 19596830 ------------GCUCCUUUGCUCCUGCGCUCC-GCGCCAUCAUUUGCAUAUAGACAUAAUAUGCAAAUAUCUGCCGCAAUCUCACGA------UGUGUGUGC------------- ------------((......))....((((...-))))....((((((((((.......))))))))))....(((((((((....))------).)))).))------------- ( -19.90, z-score = -0.97, R) >droSec1.super_1 9014255 92 - 14215200 ------------GCGCCUUUGCUCCUGCGCUCC-GCGCCAUCAUUUGCAUAUAGACAUAAUAUGCAAAUAUCUGCCGCAAUCUCACGA------UGUGUGUGCAGAACGCU----- ------------((((..........))))...-(((.....((((((((((.......)))))))))).((((((((((((....))------).)))).))))).))).----- ( -29.90, z-score = -2.59, R) >droYak2.chr2R 3669289 96 + 21139217 --------------------GCUCCUUUGCUCCUGGGCCAUCAUUUGCAUAUAGACAUAAUAUGCAAAUAUCUGCCGCAAUCUCACGAUGUGUGUGUGUGUGCAGAGCGCAAGGAU --------------------..((((((((((...(((....((((((((((.......))))))))))....)))(((....((((.....))))....))).))))).))))). ( -32.80, z-score = -1.99, R) >droEre2.scaffold_4845 8246781 94 + 22589142 ------------UCUCCCUUGCUCCUGCGCUCC-GCGCCAUCAUUUGCAUAUAGACAUAAUAUGCAAAUAUCUGCCGCAAUCUCACGA----UGUGUGUGUGCAGAAUGCC----- ------------........((..((((((..(-((((.(((((((((((((.......)))))))))).......(........)))----)))))).))))))...)).----- ( -26.00, z-score = -2.28, R) >droAna3.scaffold_13266 7209896 69 + 19884421 ----------------------UCAAGCGCUCC---CCCAUCAUUUGCAUAUAGACAUAAUAUGCAAAUAUCUGCCGCAAUCUCACU-------CGAAAAC--------------- ----------------------....((((...---......((((((((((.......))))))))))....).))).........-------.......--------------- ( -10.62, z-score = -2.26, R) >dp4.chr3 113799 104 + 19779522 UACUCCACUUUUCCUGCUCCUCUUUUGGUCUCCUGUA-GAG-AUUUGCAUAUAGACAUAAAAUGCAAAUAUCUACCGACAUCCA---GCG-AUGUGUCUGCUCAGAAGAG------ ...................((((((((((.....(((-((.-((((((((...........)))))))).))))).(((((...---...-..))))).)).))))))))------ ( -25.60, z-score = -2.34, R) >droPer1.super_37 92767 104 + 760358 UACUCCACUUUUCCUGCUCCUCUUUUGGUCUCCUGUA-GAG-AUUUGCAUAUAGACAUAAAAUGCAAAUAUCUACCGCCAUCCA---GCG-AUGUGUCUGCUCAGAAGAG------ ...................((((((((((.....(((-((.-((((((((...........)))))))).)))))(((......---)))-........)).))))))))------ ( -24.40, z-score = -2.08, R) >droGri2.scaffold_15112 3823763 87 + 5172618 -------------CAGCUUCUGCUUUGGCUUCUGCUGUUAU-AUUUGCAUAUAGACAUCAUAUGCAAAUAUGUGGCGCAAUCUAAAGGCC-AUGUGUAUGUU-------------- -------------((((....((....))....))))..((-((((((((((.(....)))))))))))))(((((.(........))))-)).........-------------- ( -24.50, z-score = -1.21, R) >consensus _____________CUCCUUUGCUCCUGCGCUCC_GCGCCAUCAUUUGCAUAUAGACAUAAUAUGCAAAUAUCUGCCGCAAUCUCACGA____UGUGUGUGUGCAGAA_GC______ ..........................................((((((((((.......))))))))))............................................... ( -8.22 = -8.44 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:09 2011