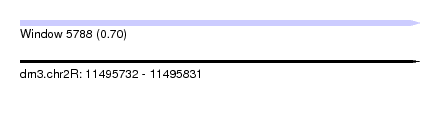
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,495,732 – 11,495,831 |
| Length | 99 |
| Max. P | 0.702670 |

| Location | 11,495,732 – 11,495,831 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 62.96 |
| Shannon entropy | 0.73795 |
| G+C content | 0.39036 |
| Mean single sequence MFE | -19.06 |
| Consensus MFE | -4.97 |
| Energy contribution | -5.12 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.702670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
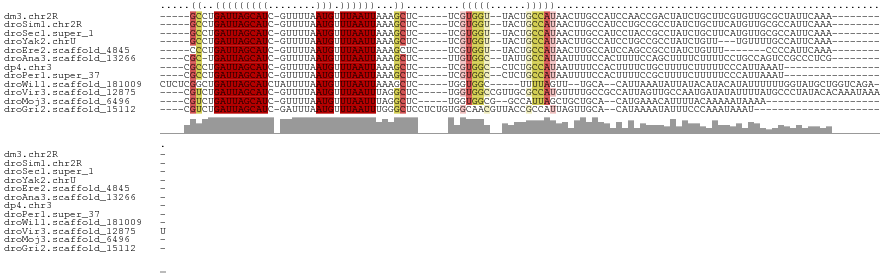
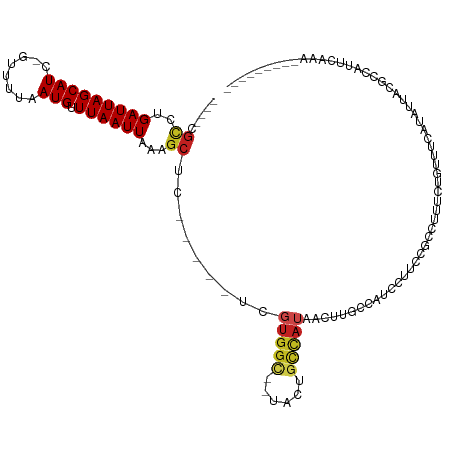
>dm3.chr2R 11495732 99 + 21146708 -----GCCUGAUUAGCAUC-GUUUUAAUGUUUAAUUAAAGCUC-----UCGUGGU--UACUGCCAUAACUUGCCAUCCAACCGACUAUCUGCUUCGUGUUGCGCUAUUCAAA--------- -----...(((.((((...-((((((((.....))))))))..-----..(((((--....)))))...........(((((((.........))).)))).)))).)))..--------- ( -22.00, z-score = -2.34, R) >droSim1.chr2R 10224374 99 + 19596830 -----GCCUGAUUAGCAUC-GUUUUAAUGUUUAAUUAAAGCUC-----UCGUGGU--UACUGCCAUAACUUGCCAUCCUGCCGCCUAUCUGCUUCAUGUUGCGCCAUUCAAA--------- -----((.((((.((((..-((((((((.....))))))))..-----..(((((--...((.((.....)).))....))))).....))))....)))).))........--------- ( -17.60, z-score = -0.45, R) >droSec1.super_1 8986219 99 + 14215200 -----GCCUGAUUAGCAUC-GUUUUAAUGUUUAAUUAAAGCUC-----UCGUGGU--UACUGCCAUAACUUGCCAUCCUACCGCCUAUCUGCUUCAUGUUGCGCCAUUCAAA--------- -----((.((((.((((..-((((((((.....))))))))..-----..(((((--...((.((.....)).))....))))).....))))....)))).))........--------- ( -17.30, z-score = -0.90, R) >droYak2.chrU 26569106 96 + 28119190 -----GCCUGAUUAGCAUC-GUUUUAAUGUUUAAUUAAAGCUC-----UCGUGGU--UACUGCCAUAACUUGCCAUCCUGCCGCCUAUCUGUU---UGUUUUGCCAUUCAAA--------- -----((..(((.((((..-((((((((.....))))))))..-----..(((((--...((.((.....)).))....))))).....))))---.)))..))........--------- ( -17.00, z-score = -1.16, R) >droEre2.scaffold_4845 8218530 92 - 22589142 -----CCCUGAUUAGCAUC-GUUUUAAUGUUUAAUUAAAGCUC-----UCGUGGU--UACUGCCAUAACUUGCCAUCCAGCCGCCUAUCUGUUU-------CCCCAUUCAAA--------- -----....(((..((...-((((((((.....))))))))..-----..(((((--....)))))................))..))).....-------...........--------- ( -14.90, z-score = -1.79, R) >droAna3.scaffold_13266 7185562 99 - 19884421 ----CGC-UGAUUAGCAUC-GUUUUAAUGUUUAAUUAAAGCUC-----UUGUGGC--UAUUGCCAUAAUUUUCCACUUUUCCAGCUUUUCUUUUCCUGCCAGUCCGCCCUCG--------- ----.((-.((((.(((..-((((((((.....))))))))..-----(((((((--....)))))))............................))).)))).)).....--------- ( -22.70, z-score = -4.26, R) >dp4.chr3 88167 92 - 19779522 ----CGCCUGAUUAGCAUC-GUUUUAAUGUUUAAUUAAAGCUC-----UCGUGGC--CUCUGCCAUAAUUUUCCACUUUUCUGCUUUUCUUUUUCCCAUUAAAU----------------- ----.....((..((((..-((((((((.....))))))))..-----..(((((--....)))))...............))))..))...............----------------- ( -17.40, z-score = -3.61, R) >droPer1.super_37 67325 92 - 760358 ----CGCCUGAUUAGCAUC-GUUUUAAUGUUUAAUUAAAGCUC-----UCGUGGC--CUCUGCCAUAAUUUUCCACUUUUCCGCUUUUCUUUUUCCCAUUAAAU----------------- ----.....((..(((...-((((((((.....))))))))..-----..(((((--....)))))................)))..))...............----------------- ( -17.10, z-score = -3.61, R) >droWil1.scaffold_181009 1968601 105 - 3585778 CUCUCGGCUGAUUAGCAUCUAUUUUAAUGUUUAAUUAAAGCUC-----UGGUGGC-----UUUUAGUU--UGCA--CAUUAAAUAUUAUACAUACAUAUUUUUUGGUAUGCUGGUCAGA-- .......((((((((((((((.((((((((..((((((((((.-----....)))-----.)))))))--...)--)))))))....................))).))))))))))).-- ( -25.50, z-score = -2.67, R) >droVir3.scaffold_12875 14103972 111 - 20611582 ----CGUCUGAUUAGCAUC-GUUUUAAUGUUUAAUUUAGGCUC-----UGGUGGCCGUUGCGCCAUGUUUUGCCGCCAUUAGUUGCCAAUGAUAUAUUUUAUGCCCUAUACACAAAUAAAU ----..........((((.-.....(((((...(((..(((.(-----(((((((.((.((.....))...)).))))))))..))))))...)))))..))))................. ( -20.80, z-score = 0.74, R) >droMoj3.scaffold_6496 23509426 87 + 26866924 ----CGUCUGAUUAGCAUC-GUUUUAAUGUUUAAUUUAGGCUC-----UGGUGGCG--GCCAUUAGCUGCUGCA--CAUGAAACAUUUUACAAAAAUAAAA-------------------- ----.((((((..(((((.-......)))))....))))))((-----(((..(((--((.....)))))..).--)).))....................-------------------- ( -20.30, z-score = -1.22, R) >droGri2.scaffold_15112 3775963 92 - 5172618 ----CGUCUGAUUAGCAUC-GAUUUAAUGUUUAAUUUGGGCUCCUCUGUGGCAACGUUACCGCCAUUAGUUGCA--CAUAAAAUAUUUCCCAAAUAAAU---------------------- ----(((..((((......-))))..)))....(((((((......(((((((((.............))))).--))))........)))))))....---------------------- ( -16.06, z-score = -0.42, R) >consensus ____CGCCUGAUUAGCAUC_GUUUUAAUGUUUAAUUAAAGCUC_____UCGUGGC__UACUGCCAUAACUUGCCAUCCUUCCGCCUUUCUGUUUCAUAUUACGCCAUUCAAA_________ .....((..(((((((((........))).))))))...)).........(((((......)))))....................................................... ( -4.97 = -5.12 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:05 2011