| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,467,951 – 11,468,045 |
| Length | 94 |
| Max. P | 0.987707 |

| Location | 11,467,951 – 11,468,045 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.89 |
| Shannon entropy | 0.63128 |
| G+C content | 0.52233 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -17.79 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.987707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
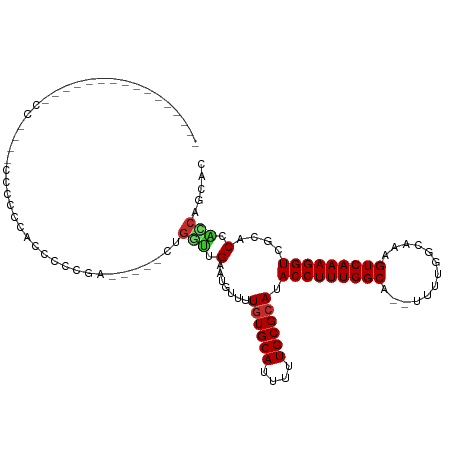
>dm3.chr2R 11467951 94 - 21146708 ----------------CCCCCCCCCCCCCCCCCCGA-----CUGGUUGAAUGUUUUGUGCAUUUUUGCGCAUACCUUUGGCA--UUUUGGCAAAGUCAAAGGUCGCACCACCAGCAC ----------------..................(.-----(((((.(.......((((((....)))))).(((((((((.--..........)))))))))....).)))))).. ( -24.70, z-score = -1.66, R) >droPer1.super_37 39905 111 + 760358 GUGCCACUGCAUGUCUCCGUCUCCUUUCCCCUCUGACUGUUCUGUUUGAGUGUGUUGUGCAUUUUUGCGCAUACCUUUGGCA--UUUUGGCAAAGUCAAAGGUCGCCCAGCCC---- ((((....)))).................(((.(((((....(((..((((((((((((((....)))))).)).....)))--)))..))).))))).)))...........---- ( -25.50, z-score = -0.29, R) >droAna3.scaffold_13266 7156375 79 + 19884421 -------------------------------GUCGA-----CUGGUUGAAUGUUUUGUGCAUUUUUGCGCAUACCUUUGGCA--UUUUGGCAAAGUCAAAGGUCGUCCCCUCGGCCG -------------------------------(((((-----..((.(((......((((((....))))))..((((((((.--..........))))))))))).))..))))).. ( -26.30, z-score = -2.42, R) >droEre2.scaffold_4845 8188779 108 + 22589142 --GCCCCCACAGCCCACCUUAACCCCCAACCCCCGA-----CUGGUUGAAUGUUUUGUGCAUUUUUGCGCAUACCUUUGGCA--UUUUGGCAAAGUCAAAGGUCGCACCACCACCAC --((..((...(((............(((((.....-----..))))).......((((((....)))))).......))).--.((((((...))))))))..))........... ( -25.00, z-score = -1.05, R) >droYak2.chr2R 3614785 93 + 21139217 -----------GCCC-CCAAAACC-----CCCCCGA-----CAGGUUGAAUGUUUUGUGCAUUUUUGCGCAUACCUUUGGCA--UUUUGGCAAAGUCAAAGGUCGCACCACCACCAC -----------....-........-----....(((-----(..((..((.((..((((((....)))))).)).))..)).--(((((((...)))))))))))............ ( -21.60, z-score = -0.40, R) >droSec1.super_1 8958904 86 - 14215200 ------------------------CCCCAUCCCCGC-----CUGGUUGAAUGUUUUGUGCAUUUUUGCGCAUACCUUUGGCA--UUUUGGCAAAGUCAAAGGUCGCACCACCAGCAC ------------------------..........((-----.((((.(.......((((((....)))))).(((((((((.--..........)))))))))....).)))))).. ( -24.70, z-score = -1.29, R) >droSim1.chr2R 10197096 86 - 19596830 ------------------------CCCCAUCCCCGC-----CUGGUUGAAUGUUUUGUGCAUUUUUGCGCAUACCUUUGGCA--UUUUGGCAAAGUCAAAGGUCGCACCACCAGCAC ------------------------..........((-----.((((.(.......((((((....)))))).(((((((((.--..........)))))))))....).)))))).. ( -24.70, z-score = -1.29, R) >droGri2.scaffold_15112 3732144 101 + 5172618 ---------------ACCCCUGCUCAAAACCAACUUCACCCCA-CUUGUUUUGUGUGUGCAUUUUUGCGCAUACCUUUGGCAGUUUUUGGCAAAGUCAAAGGUCCUGCAACUCCCCC ---------------.....(((.....(((............-........(((((((((....)))))))))(((((.(((...))).))))).....)))...)))........ ( -23.30, z-score = -1.19, R) >droMoj3.scaffold_6496 23457700 95 - 26866924 ---------------GCCCCAGCCAAACCCCAACCGAGCCCCAGCCUGGUUU---UCUGCAUUUUUGCGCAUACCUUUGGCAGUUUUUGGCAAAGUCAAAGGUCUGCCCGGCC---- ---------------......(((.......(((((.((....)).))))).---...(((....)))(((.(((((((((.((.....))...))))))))).)))..))).---- ( -28.00, z-score = -0.82, R) >consensus ________________CC____CCCCCCACCCCCGA_____CUGGUUGAAUGUUUUGUGCAUUUUUGCGCAUACCUUUGGCA__UUUUGGCAAAGUCAAAGGUCGCACCACCAGCAC ...........................................(((.(.......((((((....)))))).(((((((((.............)))))))))....).)))..... (-17.79 = -17.90 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:25:00 2011