
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,335,889 – 11,335,979 |
| Length | 90 |
| Max. P | 0.570722 |

| Location | 11,335,889 – 11,335,979 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 54.50 |
| Shannon entropy | 0.83156 |
| G+C content | 0.52622 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -8.07 |
| Energy contribution | -7.04 |
| Covariance contribution | -1.03 |
| Combinations/Pair | 2.14 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11335889 90 - 21146708 --CUGGGCCGUCCGAUAAGUUUAAUCGGCU-CAGGCUGA-AACUC--GUUUUCGGAA--UUUGGAGGUCACACUUCGAAGACCUUAACCA-U-UAGCCAG----------------- --.(((((((...((.....))...)))))-))((((((-.....--(((...((..--(((((((......)))))))..))..)))..-)-)))))..----------------- ( -27.30, z-score = -1.84, R) >droSim1.chr2R 10062312 90 - 19596830 --CUGGGCCGUCCGAUAAGUUUAAUCGGCU-CAGGCUGA-AACUC--GUUUUCGGAA--UUUGGAGGCCACACUUCGAAGACCUUAACCA-U-UUGCCAG----------------- --.(((((((...((.....))...)))))-))(((.((-((...--..))))((..--(((((((......)))))))..)).......-.-..)))..----------------- ( -25.10, z-score = -0.92, R) >droSec1.super_1 8815069 90 - 14215200 --CUGGGCCGUCCGAUAAGUUUAAUCGGCU-CAGGCUGA-AACUC--GUUUUCGGAA--UUUGGAGGCCACACUUCGAAGACCUUAACCA-U-UUGCCAG----------------- --.(((((((...((.....))...)))))-))(((.((-((...--..))))((..--(((((((......)))))))..)).......-.-..)))..----------------- ( -25.10, z-score = -0.92, R) >droYak2.chr2R 3475441 93 + 21139217 --CUGUUCGGACCGAUAAGUUUAAUCGGCUCCAGGCUGA-GCCUC--GUUUUUCGAAAUUUUGGAGGCCACACUUCGAAGUGCUAAGCCA-U-UUGCCAG----------------- --(((...((((((((.......))))).))).((((.(-((.((--(.....))).(((((((((......)))))))))))).)))).-.-....)))----------------- ( -30.20, z-score = -2.22, R) >droEre2.scaffold_4845 8051061 90 + 22589142 --CUGGGCCGACCGAUAAGUUUAAUCGGCU-CAGGCUGA-ACCUC--GUUUUCGGAA--UUCGGAGGUCACACUCUGAAAUCCUAAGCCA-U-UUGCCAG----------------- --.((((((((..((.....))..))))))-))((((((-(....--...)))(((.--(((((((......))))))).)))..)))).-.-.......----------------- ( -31.60, z-score = -2.83, R) >droAna3.scaffold_13266 7753750 85 - 19884421 GUCCGGGCGAGCUGAUAAACUUAAUCGGCCCACGGC------------UUGCCGGAA--UUUUAAGGUCCUGUAUUGGAGUCCUUAACCAGU-UCCCCAG----------------- .....(((((((((.....(......).....))))------------)))))((((--(((((((((((......)))..))))))..)))-)))....----------------- ( -28.10, z-score = -1.86, R) >dp4.chr3 11971691 113 - 19779522 --CCGGGCCAGCUGAUAAGCUUAAUCGGCUCACGCUCAACGCCUG--CCUGGUCGGGUCGCAUGGGGUCGGGUCGACGCAGGCCAAGCCAGUAUCGGCAUCCUUAACCAUUUGCCUG --((((((.(((((((.......)))))))...((.....))..)--))))).((((.((.((((....(((((((..(.(((...))).)..)))))..))....)))).)))))) ( -39.60, z-score = 0.29, R) >droPer1.super_44 183854 113 - 614636 --CUGGGCCAGCUGAUAAGCUUAAUCUGCUCACGCUCAACGCCUU--CCUGGUCGGGUCGCAUUGGGUCGGGUCGACGCAGACCAAGCCAGUAUCGGCAUCCUUAACCAUUUGCCUG --..((((..(((((((.((((..(((((((.(((((((.((..(--((.....)))..)).))))).))....)).)))))..))))...)))))))(....)........)))). ( -35.50, z-score = -0.22, R) >droWil1.scaffold_181141 2240236 87 + 5303230 --CUGGCCAGGUC--UAGGUCCAA---GUCCAGGCCCGACGUUCUGGCCUGGUUGUUUACCUUAUAUUCAUGCCAAAAGGCAGGCAACUGAUAA----------------------- --...(((.((..--.((((....---..(((((((.(.....).)))))))......)))).....)).((((....))))))).........----------------------- ( -29.60, z-score = -1.45, R) >consensus __CUGGGCCGGCCGAUAAGUUUAAUCGGCUCCAGGCUGA_ACCUC__GUUUUCGGAA__UUUGGAGGUCACACUUCGAAGACCUAAACCA_U_UUGCCAG_________________ ....((.....(((((.......)))))...............................(((((((......)))))))........))............................ ( -8.07 = -7.04 + -1.03)

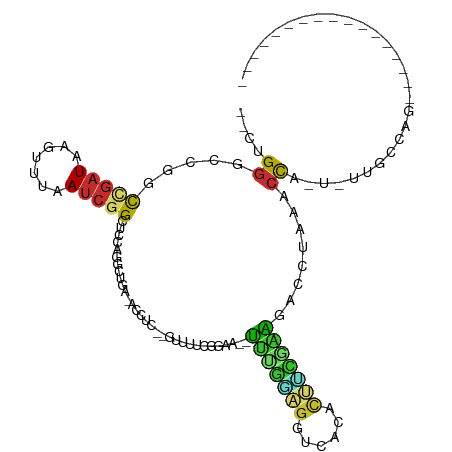
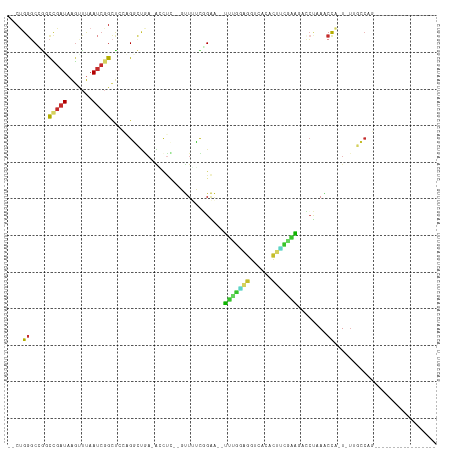
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:51 2011