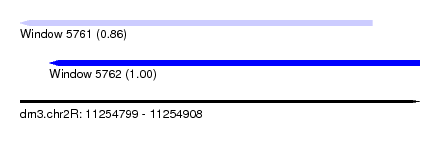
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,254,799 – 11,254,908 |
| Length | 109 |
| Max. P | 0.999034 |

| Location | 11,254,799 – 11,254,895 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 71.33 |
| Shannon entropy | 0.54128 |
| G+C content | 0.35189 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -9.04 |
| Energy contribution | -8.68 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
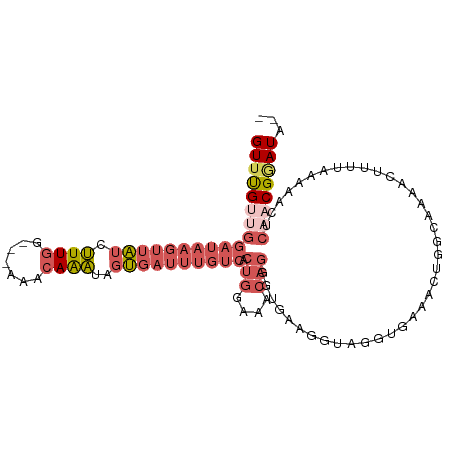
>dm3.chr2R 11254799 96 - 21146708 -GUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGUUAGGUAAAACUGGCAAAACUUUUAAAAACUC----AACGGAUACAUUGAAC----- -......((((....))))(((((((((((..(((....)))(((...(((((......)))))...)))...........----.)))))).)))))...----- ( -21.60, z-score = -2.40, R) >droEre2.scaffold_4845 7972122 95 + 22589142 -GUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGAUAGGUGAAACUGGGAAAACUUACAA----------AACGAAUACGUUCAACGGAUA -..((((((((....)))...(((.(((((..(((....)))(((.....(((......))).....))).....----------.)))))))).......))))) ( -17.50, z-score = -0.74, R) >droYak2.chr2R 3393567 100 + 21139217 -GUCAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACGGGGUGAAGAUAGGUGAAGUUGGGUGAAACUGGGAAAACUUUCAAAACGGAUACAUUAAAC----- -.(((((((((....))))......((..((.(((....)))))..))....))))).(((..(((...(((.(((....)))....)))...)))..)))----- ( -21.40, z-score = -1.44, R) >droSec1.super_1 8738785 96 - 14215200 -GUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGGUAGGUGAAACUGGCAAAACUUUUAAAAACUC----AACGGAUACAUUGAAC----- -......((((....))))(((((((((((..(((....)))(((...(.(((......))).)...)))...........----.)))))).)))))...----- ( -19.60, z-score = -1.31, R) >droSim1.chr2R 9983401 96 - 19596830 -GUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGGUAGGUGAAACUGGCAAAACUUUUAAAAACUC----AACGGAUACAUUGAAC----- -......((((....))))(((((((((((..(((....)))(((...(.(((......))).)...)))...........----.)))))).)))))...----- ( -19.60, z-score = -1.31, R) >droMoj3.scaffold_6496 24235948 100 + 26866924 GGUUAGUUUGAAAAGU---UGACAAGCUAACACAAAAAACA-UGUGAAUUUAAAUAAAAACGAAAUACUUCUCAA--ACUAACCUCGCAUAUACAUAGUAGUUAAA (((((((((((..(((---(....))))..((((.......-)))).........................))))--)))))))...................... ( -19.20, z-score = -3.06, R) >consensus _GUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGAUAGGUGAAACUGGCAAAACUUUUAAAAACUC____AACGGAUACAUUGAAC_____ ..(((((((((....)))...........((((((....)..)))))....))))))................................................. ( -9.04 = -8.68 + -0.36)
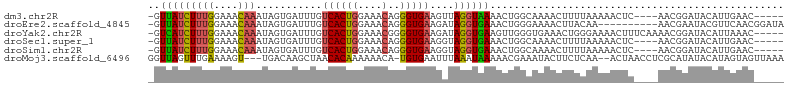
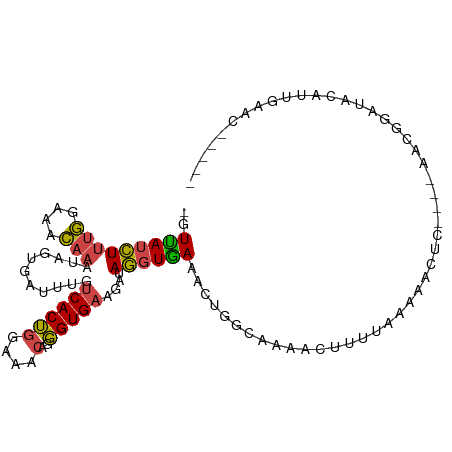
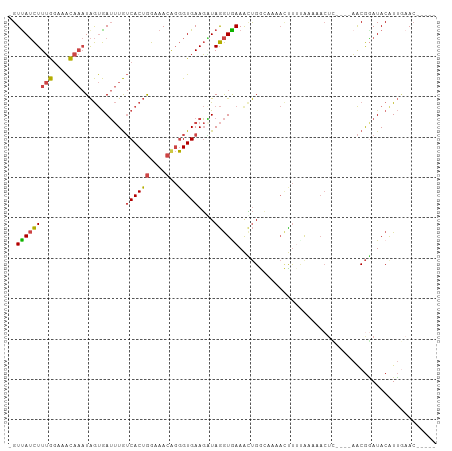
| Location | 11,254,807 – 11,254,908 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.05 |
| Shannon entropy | 0.44331 |
| G+C content | 0.34414 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -11.94 |
| Energy contribution | -12.82 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.33 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11254807 101 - 21146708 GUUUGUUGGAUUAGUUAUCUUUGG---AAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGUUAGGUAAAACUGGCAAAACUUUUAAAAACUCAACGGAUA-- (((((((((((.((((((.((((.---...))))..)))))).))).(((....)))(((...(((((......)))))...)))..........)))))))).-- ( -27.20, z-score = -3.46, R) >droEre2.scaffold_4845 7972135 95 + 22589142 GUUUGUUGGAUAAGUUAUCUUUGG---AAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGAUAGGUGAAACUGGGAAAACUUACAAAA------CGAAUA-- (((((((...((((((.((((((.---...))).((((.(((((((.(((....)))......)))))))...))))))).))))))...))------))))).-- ( -25.50, z-score = -4.03, R) >droSec1.super_1 8738793 101 - 14215200 GUUUGUUGGAUAAGUUAUCUUUGG---AAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGGUAGGUGAAACUGGCAAAACUUUUAAAAACUCAACGGAUA-- ((((((((((((((((((.((((.---...))))..)))))))))).(((....)))(((...(.(((......))).)...)))..........)))))))).-- ( -28.80, z-score = -3.94, R) >droSim1.chr2R 9983409 101 - 19596830 GUUUGUUGGAUAAGUUAUCUUUGG---AAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGGUAGGUGAAACUGGCAAAACUUUUAAAAACUCAACGGAUA-- ((((((((((((((((((.((((.---...))))..)))))))))).(((....)))(((...(.(((......))).)...)))..........)))))))).-- ( -28.80, z-score = -3.94, R) >droMoj3.scaffold_6496 24235961 105 + 26866924 GUUCGUUGAGCAAGACGAGGUUAGUUUGAAAAGUUGACAAGCUAACACAAAAAACA-UGUGAAUUUAAAUAAAAACGAAAUACUUCUCAAACUAACCUCGCAUAUA ((((...))))..(.((((((((((((((..((((....))))..((((.......-)))).........................)))))))))))))))..... ( -27.90, z-score = -4.67, R) >consensus GUUUGUUGGAUAAGUUAUCUUUGG___AAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGGUAGGUGAAACUGGCAAAACUUUUAAAAACUCAACGGAUA__ .((((((((...(.(((((((((......((((((....))))))..(((....)))..))))))))).)....))))))))........................ (-11.94 = -12.82 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:43 2011