| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,249,205 – 11,249,277 |
| Length | 72 |
| Max. P | 0.862241 |

| Location | 11,249,205 – 11,249,277 |
|---|---|
| Length | 72 |
| Sequences | 11 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 58.66 |
| Shannon entropy | 0.95513 |
| G+C content | 0.45402 |
| Mean single sequence MFE | -14.95 |
| Consensus MFE | -5.59 |
| Energy contribution | -5.82 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
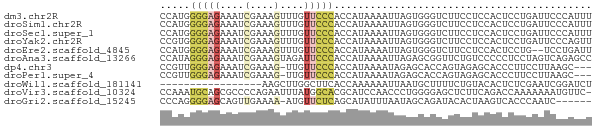
>dm3.chr2R 11249205 72 - 21146708 CCAUGGGGAGAAAUCGAAAGUUUGUUCCCACCAUAAAAUUAGUGGGUCUUCCUCCACUCCUGAUUCCCAUUU ...((((((.(((.(....)))).))))))......((((((.((((........))))))))))....... ( -18.10, z-score = -0.94, R) >droSim1.chr2R 9977915 72 - 19596830 CCAUGGGGAGAAAUCGAAAGUUUGUUCCCACCAUAAAAUUAGUGGGUCUUCCUCCACUCCUGAUUCCCAUUU ...((((((.(((.(....)))).))))))......((((((.((((........))))))))))....... ( -18.10, z-score = -0.94, R) >droSec1.super_1 8733245 72 - 14215200 CCAUGGGGAGAAAUCGAAAGUUUGUUCCCACCAUAAAAUUAGUGGGUCUUCCUCCACUCCUGAUUCCCAUUU ...((((((.(((.(....)))).))))))......((((((.((((........))))))))))....... ( -18.10, z-score = -0.94, R) >droYak2.chr2R 3387898 72 + 21139217 CCGUGGGGAGAAAUCGAAAGUUUGUUCCCACCAUAAAAUUAGUGGGUCUUCCUCCACUCCUGAUUCCCAGUU ..(((((((.(((.(....)))).))))))).........((((((......)))))).(((.....))).. ( -21.90, z-score = -2.20, R) >droEre2.scaffold_4845 7966568 70 + 22589142 CCAUGGGGAGAAAUCGAAAGUUUGUUCCCACCAUAAAAUUAGUGGGUCUUCCUCCACUCCUG--UCCUGAUU ...((((((.(((.(....)))).))))))..........((((((......))))))....--........ ( -17.90, z-score = -0.83, R) >droAna3.scaffold_13266 7670148 72 - 19884421 CCAUAGGGAGAAAUCGAAAGUAGAUUCCCACCAUAAAAUUAGAGCGGUUCUGUCCCCCUCCUAGUCAGAGCC .....(((((....(....)....)))))................(((((((.(.........).))))))) ( -17.40, z-score = -2.20, R) >dp4.chr3 15081681 68 - 19779522 CCGUUGGGAGAAAUCGAAAG-UUGUUCCCACCAUAAAAUAGAGCACCAGUAGAGCACCCUUCCUUAAGC--- ..(.((((((....(....)-...)))))).)..........((.........))..............--- ( -11.00, z-score = -0.57, R) >droPer1.super_4 3193280 68 + 7162766 CCGUUGGGAGAAAUCGAAAG-UUGUUCCCACCAUAAAAUAGAGCACCAGUAGAGCACCCUUCCUUAAGC--- ..(.((((((....(....)-...)))))).)..........((.........))..............--- ( -11.00, z-score = -0.57, R) >droWil1.scaffold_181141 2125097 55 + 5303230 -----------------AAGCUUGGCUUCACCAAAAAAUUAAUGCUUUUCUGUACACUCUCGAAUCGGAUCU -----------------((((((((.....)))).........)))).((((.............))))... ( -3.42, z-score = 1.10, R) >droVir3.scaffold_10324 499928 71 + 1288806 CCAAAUGCAGCGCCCCAGAAUUUAUGGCACGCAUCCAACCCUGGGGAGCUCUUCAGACCAAAAAAAUGUUC- ........(((.((((((......(((.......)))...)))))).))).....................- ( -15.00, z-score = -0.13, R) >droGri2.scaffold_15245 9838310 65 + 18325388 CCCAGGGGAGCAGUUGAAAA-AUGUUCUCAGCAUAUUUAAUAGCAGAUACACUAAGUCACCCAAUC------ ....(((..((.((((((..-((((.....)))).)))))).)).(((.......))).)))....------ ( -12.50, z-score = -1.39, R) >consensus CCAUGGGGAGAAAUCGAAAGUUUGUUCCCACCAUAAAAUUAGUGGGUCUUCCUCCACUCCUGAUUCCGAUUU .....(((((....(....)....)))))........................................... ( -5.59 = -5.82 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:41 2011