
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,218,249 – 11,218,386 |
| Length | 137 |
| Max. P | 0.972725 |

| Location | 11,218,249 – 11,218,386 |
|---|---|
| Length | 137 |
| Sequences | 6 |
| Columns | 139 |
| Reading direction | forward |
| Mean pairwise identity | 71.10 |
| Shannon entropy | 0.56357 |
| G+C content | 0.55640 |
| Mean single sequence MFE | -50.02 |
| Consensus MFE | -30.52 |
| Energy contribution | -31.53 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
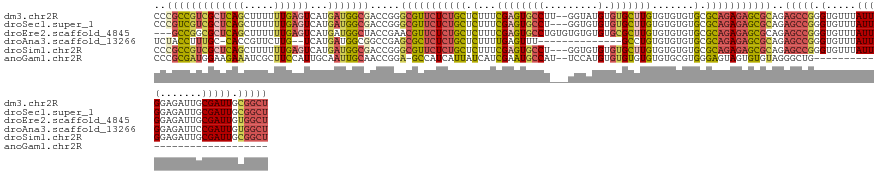
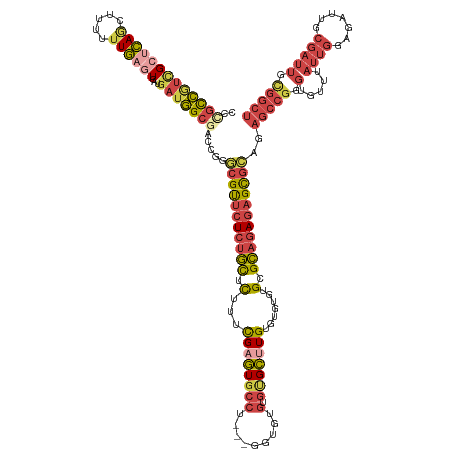
>dm3.chr2R 11218249 137 + 21146708 CCCGCCGUCGCUCAGCUUUUUUGAGUCAUGAUGGCGACCGGGCGUUCUCUGCUCUUUCGAGUGCCUU--GGUAUGUGUGCUUGUGUGUGUGCGCAGAGAGCGCAGAGCCGGGUGUUUAUUGGAGAUUGCGAUUGCGGCU .((((.(((((....((((..((((.(((..((((......(((((((((((........((((...--.))))(((..(......)..))))))))))))))...)))).)))))))..))))...))))).)))).. ( -56.70, z-score = -1.99, R) >droSec1.super_1 8710900 136 + 14215200 CCCGUCGUCGCUCAGCUUUUUUGAGUCAUGAUGGCGACCGGGCGUUCUCUGCUCUUUCGAGUGCCU---GGUGUGUGUGCUUGUGUGUGUGCGCAGAGAGCGCAGAGCCGGGUGUUUAUUGGAGAUUGCGAUUGCGGCU .((((.(((((....((((..((((.(((..((((.(((((((((((...........))))))))---))).(.(((((((.((((....))))..))))))).))))).)))))))..))))...))))).)))).. ( -55.40, z-score = -1.72, R) >droEre2.scaffold_4845 7942525 136 - 22589142 ---GCCGGCGCUCAGCUUUUUUGAGUCAUGAUGGCUACCGAACGUUCUCUGCUCUUUCGAGUGCCUGUGUGUGUGUGCGCUUGUGUGUGUGCGCAGAGAGCGCAGAGCCGGGUGUUUAUUGGAGAUUGCGAUUGUGGCU ---((((.(((....((((..((((.(((..(((((......((((((((((((....))))........(((((..(((....)))..)))))))))))))...))))).)))))))..))))...)))....)))). ( -49.10, z-score = -0.27, R) >droAna3.scaffold_13266 7635357 122 + 19884421 UCUACCUUUGC-CACCGUUCUUG--UCAUGAUGGCGGCCGAGCGCUCUCUGCUCUUUUGAGUUU--------------GCCUGUGUGUGUGCGCAGAGAGCGCAGAGCCGGGUGUUUAUUGGAGAUUCCGAUUGUGGCU .........((-(((((..(..(--((.....)))((((..(((((((((((((....))....--------------(((.......).))))))))))))).).))))..))...(((((.....))))).))))). ( -45.80, z-score = -1.59, R) >droSim1.chr2R 9955029 136 + 19596830 CCCGCCGUCGCUCAGCUUUUUUGAGUCAUGAUGGCGACCGGGCGUUCUCUGCUCUUUCGAGUGCCU---GGUGUGUGUGCUUGUGUGUGUGCGCAGAGAGCGCAGAGCCGGGUGUUUAUUGGAGAUUGCGAUUGCGGCU .((((.(((((....((((..((((.(((..((((.(((((((((((...........))))))))---))).(.(((((((.((((....))))..))))))).))))).)))))))..))))...))))).)))).. ( -56.90, z-score = -1.87, R) >anoGam1.chr2R 16557442 107 + 62725911 CCCGCGAUGGAAGAAAUCGCUUCCAUUGCAAUUGCAACCGGA-GCCAUCAUUAUCAUCGAAUGCCAU--UCCAUGUGUGUGUGUGUGCGUGGGAGUAGUGUGUAGGGCUG----------------------------- (((((((((((((......)))))))))).(((((..(((..-((((((((...(((.((((...))--)).)))...))).))).)).)))..))))).....)))...----------------------------- ( -36.20, z-score = -1.43, R) >consensus CCCGCCGUCGCUCAGCUUUUUUGAGUCAUGAUGGCGACCGGGCGUUCUCUGCUCUUUCGAGUGCCU___GGUGUGUGUGCUUGUGUGUGUGCGCAGAGAGCGCAGAGCCGGGUGUUUAUUGGAGAUUGCGAUUGCGGCU ..(((((((((((((.....))))))...))))))).(((((((((((((((.(...((((((((.........).))))))).......).)))))))))))....))))............................ (-30.52 = -31.53 + 1.01)
| Location | 11,218,249 – 11,218,386 |
|---|---|
| Length | 137 |
| Sequences | 6 |
| Columns | 139 |
| Reading direction | reverse |
| Mean pairwise identity | 71.10 |
| Shannon entropy | 0.56357 |
| G+C content | 0.55640 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -20.20 |
| Energy contribution | -21.50 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
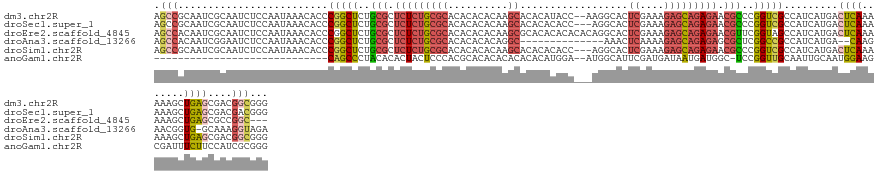
>dm3.chr2R 11218249 137 - 21146708 AGCCGCAAUCGCAAUCUCCAAUAAACACCCGGCUCUGCGCUCUCUGCGCACACACACAAGCACACAUACC--AAGGCACUCGAAAGAGCAGAGAACGCCCGGUCGCCAUCAUGACUCAAAAAAGCUGAGCGACGGCGGG ..((((..((((.................(((((..(((.(((((((((..........)).........--.......((....))))))))).)))..)))))...(((.(.((......))))))))))..)))). ( -41.60, z-score = -2.10, R) >droSec1.super_1 8710900 136 - 14215200 AGCCGCAAUCGCAAUCUCCAAUAAACACCCGGCUCUGCGCUCUCUGCGCACACACACAAGCACACACACC---AGGCACUCGAAAGAGCAGAGAACGCCCGGUCGCCAUCAUGACUCAAAAAAGCUGAGCGACGACGGG ..(((...((((.................(((((..(((.(((((((............((.(.......---.)))..((....))))))))).)))..)))))...(((.(.((......))))))))))...))). ( -36.60, z-score = -1.15, R) >droEre2.scaffold_4845 7942525 136 + 22589142 AGCCACAAUCGCAAUCUCCAAUAAACACCCGGCUCUGCGCUCUCUGCGCACACACACAAGCGCACACACACACAGGCACUCGAAAGAGCAGAGAACGUUCGGUAGCCAUCAUGACUCAAAAAAGCUGAGCGCCGGC--- .(((..........................(((((((((.(((((((((.(........).))................((....))))))))).)))..)).)))).....(.((((.......)))))...)))--- ( -34.20, z-score = -0.64, R) >droAna3.scaffold_13266 7635357 122 - 19884421 AGCCACAAUCGGAAUCUCCAAUAAACACCCGGCUCUGCGCUCUCUGCGCACACACACAGGC--------------AAACUCAAAAGAGCAGAGAGCGCUCGGCCGCCAUCAUGA--CAAGAACGGUG-GCAAAGGUAGA .(((((..((((.....))..........(((((..(((((((((((((.(.......)))--------------....((....)))))))))))))..))))).........--...))...)))-))......... ( -45.50, z-score = -3.98, R) >droSim1.chr2R 9955029 136 - 19596830 AGCCGCAAUCGCAAUCUCCAAUAAACACCCGGCUCUGCGCUCUCUGCGCACACACACAAGCACACACACC---AGGCACUCGAAAGAGCAGAGAACGCCCGGUCGCCAUCAUGACUCAAAAAAGCUGAGCGACGGCGGG ..((((..((((.................(((((..(((.(((((((............((.(.......---.)))..((....))))))))).)))..)))))...(((.(.((......))))))))))..)))). ( -41.90, z-score = -2.10, R) >anoGam1.chr2R 16557442 107 - 62725911 -----------------------------CAGCCCUACACACUACUCCCACGCACACACACACACAUGGA--AUGGCAUUCGAUGAUAAUGAUGGC-UCCGGUUGCAAUUGCAAUGGAAGCGAUUUCUUCCAUCGCGGG -----------------------------.................(((..((.((..((......))..--.))))...(((((.....(((.((-(.(.(((((....))))).).))).))).....))))).))) ( -24.80, z-score = -0.27, R) >consensus AGCCGCAAUCGCAAUCUCCAAUAAACACCCGGCUCUGCGCUCUCUGCGCACACACACAAGCACACACACC___AGGCACUCGAAAGAGCAGAGAACGCCCGGUCGCCAUCAUGACUCAAAAAAGCUGAGCGACGGCGGG .(((.........................(((((..(((.(((((((((..........))..................((....))))))))).)))..))))).........((((.......))))....)))... (-20.20 = -21.50 + 1.30)
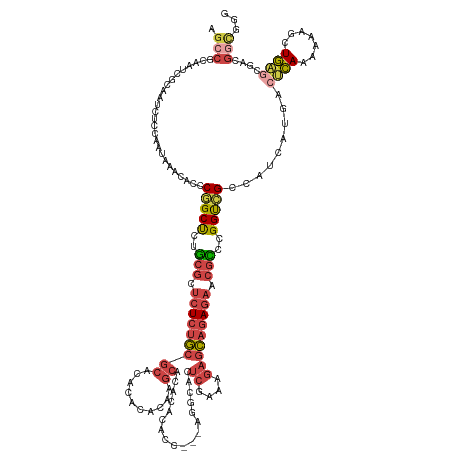
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:40 2011