
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 335,064 – 335,159 |
| Length | 95 |
| Max. P | 0.507233 |

| Location | 335,064 – 335,159 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.27 |
| Shannon entropy | 0.47169 |
| G+C content | 0.47928 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.93 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.507233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 335064 95 - 23011544 GGCCGAAUUGUCUCAUGUAGAAAGUAUUUCAAAGAU-UUU-------UCUUGG-GGCAACACGAGGCGGAUGUGCGAU-------GCAGAUUGAUUGUACAG-CAGUCGGAG ..((...((((((((...((((((.(((.....)))-)))-------))))))-))))).....(((...((((((((-------........)))))))).-..))))).. ( -24.70, z-score = -0.62, R) >droSim1.chr2L 354755 104 - 22036055 GGCCGGAUUGUCUCAUGUAGAAAGUAUUUCAAAGAUAUUU-------UCUCGG-GGCAACACGAGGCGGAUGUGUGAUGCAGGCCGCAGAUUGAUUGUACAGUCAGUCGGAG ...((..(((((((....((((((((((.....)))))))-------))).))-)))))..))..((((.(((.....)))..)))).((((((((....)))))))).... ( -35.20, z-score = -2.66, R) >droSec1.super_14 332428 104 - 2068291 GGCCGGAUUGCCUCAUGUAGAAAGUAUUUCAAAGAUAUUU-------UCUCGG-GGCAACACGAGGCGGAUGUGUGAUGCAGGCCGCAGAUUGAUUGUACAGUCAGUCGGAG ...((..(((((((....((((((((((.....)))))))-------))).))-)))))..))..((((.(((.....)))..)))).((((((((....)))))))).... ( -37.90, z-score = -3.19, R) >droYak2.chr2L 331692 103 - 22324452 GGCCUGAUUGUCUCAUGUGCAAAGUAUUUUAGAGAUACUU-------UGUCGGCGGCAACACGGGGCGGAUGUGUGAUGCAGGCUG-AGAUUGAUUGUACAGUCAGUCGGA- ((((((...(((.(((((((((((((((.....)))))))-------)))((..(....).))......))))).))).)))))).-.((((((((....))))))))...- ( -35.70, z-score = -2.05, R) >droEre2.scaffold_4929 390698 112 - 26641161 GGCCUGAUUGUCUCAUGUGGAAAGUAUUUCAGGGAUAUUUGCGGAUAUUUCGGUGGCAACACGAGGCGGAUGUGUGAUGCAGGCCGCAGAUUGAUUGUACAGUCAGUCGGAC ((((((.(((((((...(((((....)))))(..(((((....)))))..).(((....))))))))))..........))))))...((((((((....)))))))).... ( -38.00, z-score = -2.07, R) >droAna3.scaffold_12943 121486 89 + 5039921 GCUUUGAUUUUCUCUUAUA------ACUCGAAUGGCCUUUG------UUGUGGCGGC-ACACAGGGCGUAUGCGUAUUUC--------AGUUAAUGAUAU--UCAAUCGAGC ((((.((((.....(((((------(((.((((((((((.(------(.(((....)-)))))))))(....).))))).--------)))).))))...--..)))))))) ( -18.10, z-score = 0.43, R) >consensus GGCCGGAUUGUCUCAUGUAGAAAGUAUUUCAAAGAUAUUU_______UCUCGG_GGCAACACGAGGCGGAUGUGUGAUGCAGGCCGCAGAUUGAUUGUACAGUCAGUCGGAG ..((...(((((((.......(((((((.....)))))))..............(....)..)))))))...................((((((((....)))))))))).. (-14.04 = -14.93 + 0.90)

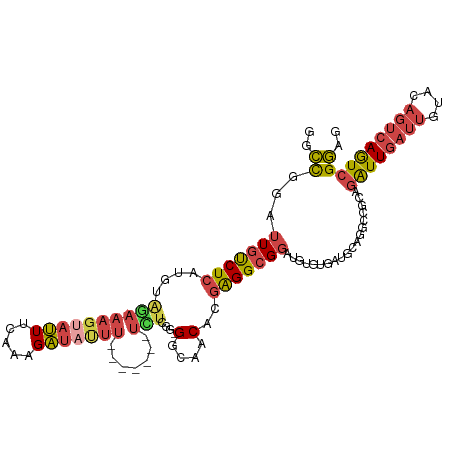

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:33 2011