| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,189,415 – 11,189,518 |
| Length | 103 |
| Max. P | 0.970092 |

| Location | 11,189,415 – 11,189,518 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.00 |
| Shannon entropy | 0.50873 |
| G+C content | 0.40827 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -10.01 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
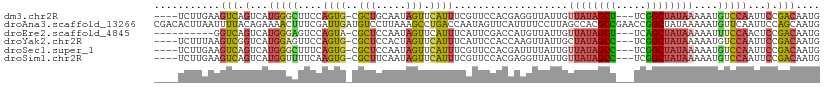
>dm3.chr2R 11189415 103 + 21146708 ----UCUUGAAGUCAGUCAUGGGCUUCCAGUG-CGCUGCAAUAGUUCAUUUCGUUCCACGAGGUUAUUGUUAUAGCU---UCGGCUAUAAAAAUGUCCAAUUCCGACAAUG ----...........((((((((((..(((..-..)))....))))))).(((.....)))((.((((.(((((((.---...))))))).)))).))......))).... ( -27.30, z-score = -2.40, R) >droAna3.scaffold_13266 7608753 111 + 19884421 CGACACUUAAUUUUACAGAAAACUUUCGAUUGAUGUCCUUAAAGCCUGACCAAUAGUUCAUUUUCCUUAGCCACGCCGAACCGGCUAUAAAAAUGUUCAAUUCCAGCAAUG .((((.((((((....((....))...))))))))))......((.((...(((.(..((((((...(((((..........)))))..))))))..).))).)))).... ( -15.90, z-score = -0.79, R) >droEre2.scaffold_4845 7913045 97 - 22589142 ----------GGUCAGUCAUGGGAGUCCAGUA-CGCUCCAAUAGUUCAUUUCAUUCGACCAUGUUAUUGUUAUAGCU---UCAGCUAUAAAAAUUUCCAACUCCGACAAUG ----------.(((...(((((((((......-.)))))...........((....)).))))......(((((((.---...)))))))..............))).... ( -18.10, z-score = -1.44, R) >droYak2.chr2R 11117215 103 + 21139217 ----UCUUUAAGUCGGUCAUGGAGUUCCAGUG-CGCUCCACUAGUUCAUUUCAUUCCACCAAGUUAUUGCUAUAGCC---UCGGCUAUAAAAAUGUCCAAUUCCGACAAUG ----.......(((((...(((((..(.((((-.....)))).)..).................((((..((((((.---...))))))..))))))))...))))).... ( -24.90, z-score = -3.39, R) >droSec1.super_1 8681921 103 + 14215200 ----UCUUGAAGUCAGUCAUGGGCUUUCAGUG-CGCUCCAAUAGUUCAUUUCGUUCCACGAUUUUAUUGUUAUAGCC---UCGGCUAUAAAAAUGUCCAAUUCCGACAAUG ----.......(((.(...(((((........-.....((((((......(((.....)))..))))))(((((((.---...)))))))....)))))...).))).... ( -21.70, z-score = -1.67, R) >droSim1.chr2R 9926393 103 + 19596830 ----UCUUGAAGUCAGUCAUGGUUUUCAAGUG-CGCUUCAAUAGUUCAUUUCGUUCCACGAGGUUAUUGUUAUAGCC---UCGGCUAUAAAAAUGUCCAAUUCCGACAAUG ----.......(((..((.(((.....(((((-.(((.....))).)))))....))).))((.((((.(((((((.---...))))))).)))).))......))).... ( -24.70, z-score = -2.85, R) >consensus ____UCUUGAAGUCAGUCAUGGGCUUCCAGUG_CGCUCCAAUAGUUCAUUUCAUUCCACGAUGUUAUUGUUAUAGCC___UCGGCUAUAAAAAUGUCCAAUUCCGACAAUG ...........(((.(...(((((..........(((.....)))........................((((((((.....))))))))....)))))...).))).... (-10.01 = -10.60 + 0.59)
| Location | 11,189,415 – 11,189,518 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.00 |
| Shannon entropy | 0.50873 |
| G+C content | 0.40827 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -15.86 |
| Energy contribution | -15.62 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
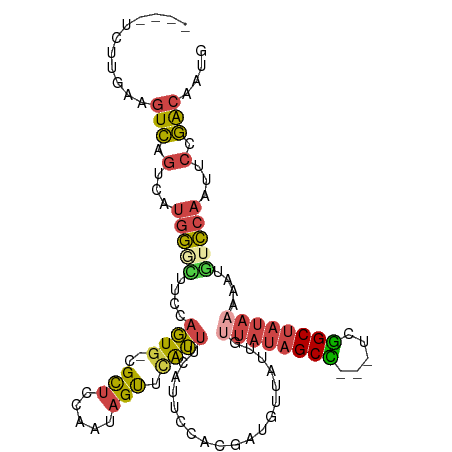
>dm3.chr2R 11189415 103 - 21146708 CAUUGUCGGAAUUGGACAUUUUUAUAGCCGA---AGCUAUAACAAUAACCUCGUGGAACGAAAUGAACUAUUGCAGCG-CACUGGAAGCCCAUGACUGACUUCAAGA---- ....(((((...(((.((((((((((((...---.)))))).......((....))...))))))..(((.(((...)-)).)))....)))...))))).......---- ( -21.90, z-score = -0.99, R) >droAna3.scaffold_13266 7608753 111 - 19884421 CAUUGCUGGAAUUGAACAUUUUUAUAGCCGGUUCGGCGUGGCUAAGGAAAAUGAACUAUUGGUCAGGCUUUAAGGACAUCAAUCGAAAGUUUUCUGUAAAAUUAAGUGUCG ....(((((...((((....))))...)))))..(((.(((((((((........)).))))))).))).....(((((........((....))..........))))). ( -20.27, z-score = 0.69, R) >droEre2.scaffold_4845 7913045 97 + 22589142 CAUUGUCGGAGUUGGAAAUUUUUAUAGCUGA---AGCUAUAACAAUAACAUGGUCGAAUGAAAUGAACUAUUGGAGCG-UACUGGACUCCCAUGACUGACC---------- ....(((((((((.(..(((.(((((((...---.))))))).)))...(((.((.((((........)))).)).))-)..).))))))...))).....---------- ( -22.70, z-score = -1.88, R) >droYak2.chr2R 11117215 103 - 21139217 CAUUGUCGGAAUUGGACAUUUUUAUAGCCGA---GGCUAUAGCAAUAACUUGGUGGAAUGAAAUGAACUAGUGGAGCG-CACUGGAACUCCAUGACCGACUUAAAGA---- ....(((((...((((((((((....(((((---(..(((....))).)))))).....))))))..((((((.....-))))))...))))...))))).......---- ( -31.60, z-score = -3.84, R) >droSec1.super_1 8681921 103 - 14215200 CAUUGUCGGAAUUGGACAUUUUUAUAGCCGA---GGCUAUAACAAUAAAAUCGUGGAACGAAAUGAACUAUUGGAGCG-CACUGAAAGCCCAUGACUGACUUCAAGA---- ....(((((...(((.(....(((((((...---.)))))))(((((...((((........))))..))))).....-........).)))...))))).......---- ( -21.70, z-score = -1.09, R) >droSim1.chr2R 9926393 103 - 19596830 CAUUGUCGGAAUUGGACAUUUUUAUAGCCGA---GGCUAUAACAAUAACCUCGUGGAACGAAAUGAACUAUUGAAGCG-CACUUGAAAACCAUGACUGACUUCAAGA---- ....(((((....((..(((.(((((((...---.))))))).)))..))((((((.....((((..((.....))..-)).)).....))))))))))).......---- ( -22.40, z-score = -2.07, R) >consensus CAUUGUCGGAAUUGGACAUUUUUAUAGCCGA___GGCUAUAACAAUAACAUCGUGGAACGAAAUGAACUAUUGGAGCG_CACUGGAAACCCAUGACUGACUUCAAGA____ ....(((((...(((......((((((((.....))))))))(((((...((((........))))..)))))................)))...)))))........... (-15.86 = -15.62 + -0.24)

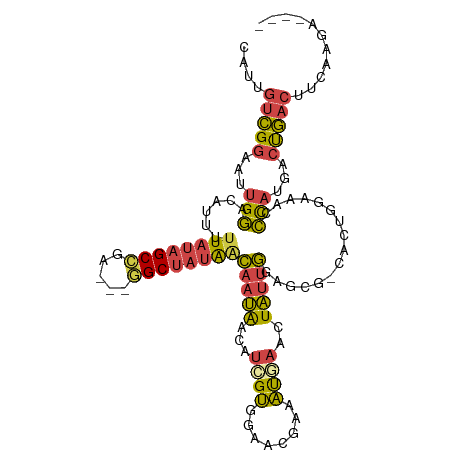
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:36 2011