| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,187,722 – 11,187,818 |
| Length | 96 |
| Max. P | 0.985874 |

| Location | 11,187,722 – 11,187,818 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.62 |
| Shannon entropy | 0.55079 |
| G+C content | 0.39352 |
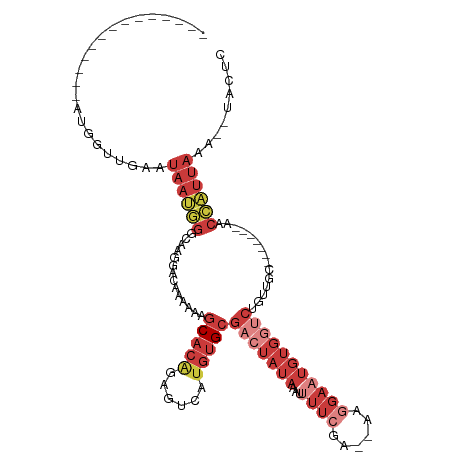
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -9.24 |
| Energy contribution | -12.30 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.985874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
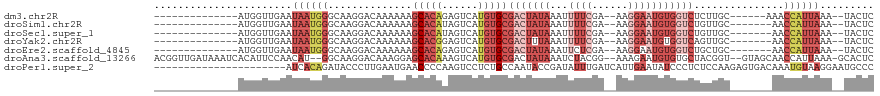
>dm3.chr2R 11187722 96 - 21146708 --------------AUGGUUGAAUAAUGGGCAAGGACAAAAAAGCACAGAGUCAUGUGCGACUAUAAAUUUUCGA--AAGGAAUGUGGUCUCUUGC------AAACCAUUAAA--UACUC --------------..(((....((((((((((((........(((((......)))))(((((((...((((..--..)))))))))))))))))------...))))))..--.))). ( -30.10, z-score = -4.63, R) >droSim1.chr2R 9924655 95 - 19596830 --------------AUGGUUGAAUAAUGGGCAAGGACAAAAAAGCACAUAGUCAUGUGCGACUAUAAAUUUUCGA--AAGGAAUGUGGUCUGUUGC-------AACCAUUAAA--UACUC --------------..(((....((((((((((.(........((((((....))))))(((((((...((((..--..)))))))))))).))))-------..))))))..--.))). ( -27.40, z-score = -3.66, R) >droSec1.super_1 8680227 95 - 14215200 --------------AUGGUUGAAUAAUGGGCAAGGACAAAAAAGCACAUAGUCAUGUGCGACUAUAAAUUUUCGA--AAGGAAUGUGGUCUGUUGC-------AACCAUUAAA--UACUC --------------..(((....((((((((((.(........((((((....))))))(((((((...((((..--..)))))))))))).))))-------..))))))..--.))). ( -27.40, z-score = -3.66, R) >droYak2.chr2R 11115456 95 - 21139217 --------------AUGGUUGAAUAAUGGGCAAGGACAAAAAAGCACGGAGUCAUGUGCGACUUUAAAUUUUCGA--AAGGAAUGUGGUCAGUUGC-------AACCAUUAAA--UACUC --------------..(((....((((((((((..........(((((......)))))((((......((((..--..))))...))))..))))-------..))))))..--.))). ( -23.50, z-score = -2.04, R) >droEre2.scaffold_4845 7911338 95 + 22589142 --------------AUGGUUGAAUAAUGGGCAAGGACAAAAAAGCACAGAGUCAUGUGCGACUAUAAAUUCUCGA--AAGGAAUGUGGUCUGCUGC-------AACCAUUAAA--UACUC --------------(((((((.......((((...........(((((......)))))((((((..((((.(..--..))))))))))))))).)-------))))))....--..... ( -27.50, z-score = -3.38, R) >droAna3.scaffold_13266 7606692 113 - 19884421 ACGGUUGAUAAAUCACAUUCCAACAU--GGCAAGGACAAAGGAGCACAAAGUCAUGUGCGACUAUAAAUCUACGG--AAAGAAUGUGUGCUACGGU--GUAGCAACCAUUAAA-GCACUC ..((((.......(((((((......--...........((..(((((......)))))..)).....((....)--)..))))))).(((((...--)))))))))......-...... ( -27.90, z-score = -1.52, R) >droPer1.super_2 740921 98 - 9036312 ----------------------AUCACAGAUACCCUUGAAUGAACCCCAAGUCCUCUGCCAAUACCGAUAUUUGAUCAUUGAAUAUCCCUCUCCAAGAGUGACAAAUGUAAGGAAUGCCC ----------------------...........(((((..((.....)).(((((((.........((((((..(...)..))))))........)))).))).....)))))....... ( -16.13, z-score = -1.23, R) >consensus ______________AUGGUUGAAUAAUGGGCAAGGACAAAAAAGCACAGAGUCAUGUGCGACUAUAAAUUUUCGA__AAGGAAUGUGGUCUGUUGC_______AACCAUUAAA__UACUC .......................((((((..............(((((......)))))(((((((...((((......)))))))))))...............))))))......... ( -9.24 = -12.30 + 3.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:35 2011