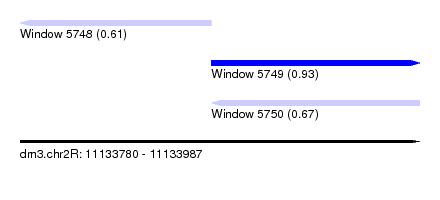
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,133,780 – 11,133,987 |
| Length | 207 |
| Max. P | 0.934390 |

| Location | 11,133,780 – 11,133,879 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 70.07 |
| Shannon entropy | 0.56098 |
| G+C content | 0.40323 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -9.76 |
| Energy contribution | -10.98 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11133780 99 - 21146708 GAGGGGUUUGUUU--GAGCUCAGUAAAAA---UAUUAUUUCCAGGCAGAGGCAGAGAAUGAACUUAUUGCACAUUAGUGGCUAUGGAAAUAGAUCCUUCGAGAG (((((((((((((--.(((.((((((...---..(((((..(..((....)).)..)))))..))))))(((....))))))....)))))))))))))..... ( -23.90, z-score = -1.02, R) >droSim1.chr2R 9871110 101 - 19596830 GAGGGUGUUGUUU--GAGCUCAGGAAAAAUAUUAUUAUUUCCAGGCAG-UGCAGCGAGUGAACUUAUUGCACAUUAGUGGCUAUAGAAAUAGAUCCUUCGAGAG (((((..((((((--.(((.(((((((.(......).))))).....(-(((((..((....))..)))))).....)))))....))))))..)))))..... ( -28.50, z-score = -2.24, R) >droSec1.super_1 8626035 101 - 14215200 GAAGGGGUUGUUU--GAGCUCAGGAAAAAUAUUAUUAUUUCCAGGCAG-UGCAGGGAAUGAACUUAUUGCCCAUUAGUGGCUAUAGAAAUAGAUCCUUCGAGAG (((((..((((((--.(((.(((((((.(......).))))).(((((-((.((........)))))))))......)))))....))))))..)))))..... ( -27.30, z-score = -1.55, R) >droYak2.chr2R 11053484 97 - 21139217 GAGGGGUUUGUUU--GAGCUCUG-GAAAG---UAAUAUUUCCAGGCAG-UGCAGGGAAUGAACGUAUGGCACAUUAGUGACUAUGGAAAUAUAUCCUUCGAGAG (((((((.(((((--..((.(((-((((.---.....))))))))).(-(.((.....)).)).((((((((....))).))))).))))).)))))))..... ( -29.60, z-score = -2.76, R) >droEre2.scaffold_4845 7852337 97 + 22589142 GAGGGGUUUGUUU--GUGCUCAG-GAAAA---CAAUAUUUCCAGGCAG-CACAGGGUAUGAACUUACUGCACAUUAGUGGCUAUGGAAAUAGAUCCUUCGAGGG (((((((((((((--((((((.(-((((.---.....)))))..).))-)))..((((......)))).(((....))).......)))))))))))))..... ( -29.20, z-score = -2.10, R) >droAna3.scaffold_13266 7555571 101 - 19884421 AAGCAGUUUACUUCAAACCCCACGAUCAAAAGUAAAAUGUACUUUUCA-GAAGAAAAAAAAAAGCAUUGAUUGUGCAAGGACAAAAUAAUUUGUUUCCCGUU-- .(((................(((((((((..(((.....)))(((((.-...))))).........)))))))))...(((((((....)))))))...)))-- ( -14.00, z-score = -0.37, R) >consensus GAGGGGUUUGUUU__GAGCUCAGGAAAAA___UAAUAUUUCCAGGCAG_UGCAGGGAAUGAACUUAUUGCACAUUAGUGGCUAUAGAAAUAGAUCCUUCGAGAG (((((((((((((...(((.........................((....)).................(((....))))))....)))))))))))))..... ( -9.76 = -10.98 + 1.23)
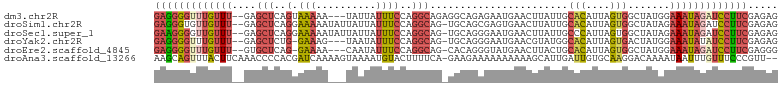
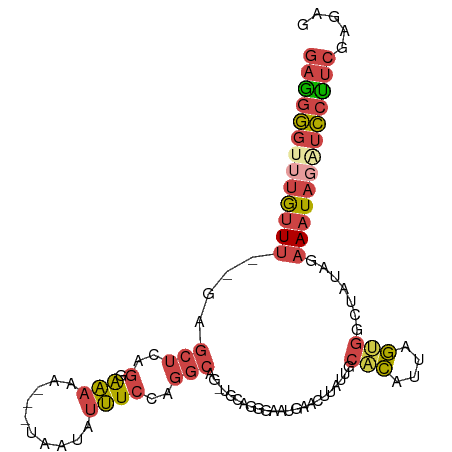
| Location | 11,133,879 – 11,133,987 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.94 |
| Shannon entropy | 0.36291 |
| G+C content | 0.36668 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -16.61 |
| Energy contribution | -17.25 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
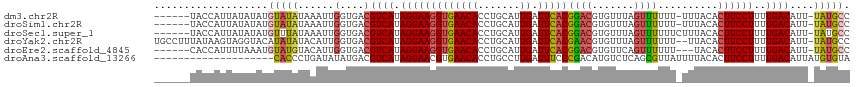
>dm3.chr2R 11133879 108 + 21146708 ------UACCAUUAUAUAUGUAUAUAAAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGGACGUGUUUAGUUUUUU-UUUACACUUCCUUUUGACAUU-UAUGCC ------(((((((((((....))))))..)))))..((((.(((((((((((.......(((((..(((....))).))))).....-))))).))))))..))))...-...... ( -25.30, z-score = -2.16, R) >droSim1.chr2R 9871211 108 + 19596830 ------UACCAUUAUAUAUGUAUAUAAAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGGACGUGUUUAGUUUUUU-UUUACACUUCCUUUUGACAUU-UAUGCC ------(((((((((((....))))))..)))))..((((.(((((((((((.......(((((..(((....))).))))).....-))))).))))))..))))...-...... ( -25.30, z-score = -2.16, R) >droSec1.super_1 8626136 109 + 14215200 ------UACCAUUAUAUAUGUUUAUAAAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGGACGUGUUUAGUUUUUUCUUUACACUUCCUUUUGACAUU-UAUGCC ------......((((.(((((.........((((.((((...(.((((....)))).)..))))))))(((.((((..((......))..)))).)))....))))).-)))).. ( -25.70, z-score = -2.23, R) >droYak2.chr2R 11053581 113 + 21139217 UGCCUUUAUAAGUAGGUACAUAUAUACAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGAACGUGUUUAGUUUUUU--UUACACUUCCUUUUGACAUU-UAUGCC (((((........)))))((((........(....)((((.(((((((((((.......(((((..(((....))).)))))....)--)))).))))))..))))...-)))).. ( -25.90, z-score = -1.64, R) >droEre2.scaffold_4845 7852434 106 - 22589142 ------CACCAUUUUAAAUGUAUGUACAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGGACGUGUUCAGUUUUUU---UACACUUCCUUUUGACAUU-UAUGCC ------(((((......((((....)))))))))..((((.((((((.((((((((((..........)))..)))))))((.....---.)).))))))..))))...-...... ( -26.90, z-score = -1.97, R) >droAna3.scaffold_13266 7555672 96 + 19884421 --------------------CACCCUGAUAUAUGACGUCAUAGGAACGUGAACACCUGCCUUGAUUUCCCGACAUGUCUCAGCGUUAUUUUACACUUCCUUUUGACAUUAUGUGUA --------------------.......((((((((.((((.(((((.(((((....(((...(((..........)))...)))....)))))..)))))..)))).)))))))). ( -20.70, z-score = -2.05, R) >consensus ______UACCAUUAUAUAUGUAUAUAAAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGGACGUGUUUAGUUUUUU_UUUACACUUCCUUUUGACAUU_UAUGCC ....................................((((.(((((((((((.......(((((..(((....))).)))))......))))).))))))..)))).......... (-16.61 = -17.25 + 0.64)
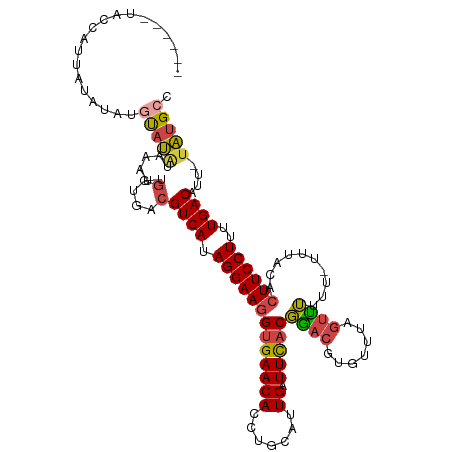
| Location | 11,133,879 – 11,133,987 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.94 |
| Shannon entropy | 0.36291 |
| G+C content | 0.36668 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.16 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.671733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
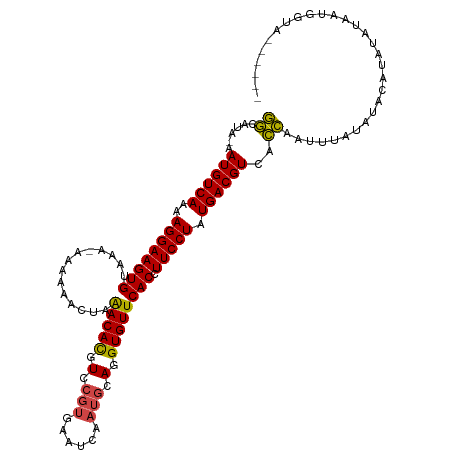
>dm3.chr2R 11133879 108 - 21146708 GGCAUA-AAUGUCAAAAGGAAGUGUAAA-AAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUUUAUAUACAUAUAUAAUGGUA------ ......-.((((((..((((((((....-.........(((((.(.(((......))).).))))))).)))))).)))))).((((..((((((....)))))))))).------ ( -22.11, z-score = -1.75, R) >droSim1.chr2R 9871211 108 - 19596830 GGCAUA-AAUGUCAAAAGGAAGUGUAAA-AAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUUUAUAUACAUAUAUAAUGGUA------ ......-.((((((..((((((((....-.........(((((.(.(((......))).).))))))).)))))).)))))).((((..((((((....)))))))))).------ ( -22.11, z-score = -1.75, R) >droSec1.super_1 8626136 109 - 14215200 GGCAUA-AAUGUCAAAAGGAAGUGUAAAGAAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUUUAUAAACAUAUAUAAUGGUA------ ......-.((((((..((((((.((..((......)).(((((.(.(((......))).).))))).)))))))).)))))).((((..(((((......))))))))).------ ( -22.00, z-score = -1.55, R) >droYak2.chr2R 11053581 113 - 21139217 GGCAUA-AAUGUCAAAAGGAAGUGUAA--AAAAAACUAAACACGUUCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUGUAUAUAUGUACCUACUUAUAAAGGCA .((...-.((((((..((((((((...--.........(((((.(.(((......))).).))))))).)))))).)))))).......((((....))))............)). ( -20.10, z-score = -0.27, R) >droEre2.scaffold_4845 7852434 106 + 22589142 GGCAUA-AAUGUCAAAAGGAAGUGUA---AAAAAACUGAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUGUACAUACAUUUAAAAUGGUG------ ......-.((((((..((((((.((.---.....))(((((((.(.(((......))).).))))))).)))))).))))))(((((((((....))))......)))))------ ( -26.50, z-score = -2.23, R) >droAna3.scaffold_13266 7555672 96 - 19884421 UACACAUAAUGUCAAAAGGAAGUGUAAAAUAACGCUGAGACAUGUCGGGAAAUCAAGGCAGGUGUUCACGUUCCUAUGACGUCAUAUAUCAGGGUG-------------------- ........((((((..(((((((((......))))((((((.((((..(....)..)))).)).))))..))))).))))))..............-------------------- ( -19.80, z-score = -0.44, R) >consensus GGCAUA_AAUGUCAAAAGGAAGUGUAAA_AAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUUUAUAUACAUAUAUAAUGGUA______ ........((((((..((((((((..............(((((.(.(((......))).).)))))))).))))).)))))).................................. (-15.94 = -16.16 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:32 2011