| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,105,810 – 11,105,861 |
| Length | 51 |
| Max. P | 0.891616 |

| Location | 11,105,810 – 11,105,861 |
|---|---|
| Length | 51 |
| Sequences | 5 |
| Columns | 56 |
| Reading direction | forward |
| Mean pairwise identity | 67.84 |
| Shannon entropy | 0.56634 |
| G+C content | 0.34412 |
| Mean single sequence MFE | -10.96 |
| Consensus MFE | -4.37 |
| Energy contribution | -5.37 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.690231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
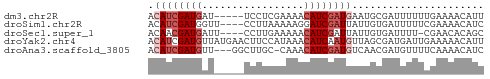
>dm3.chr2R 11105810 51 + 21146708 ACAUCGAUGAU-----UCCUCGAAAACAUCGAUGAAUGCGAUUUUUUGAAAACAUU .((((((((.(-----(......)).))))))))...................... ( -9.80, z-score = -1.24, R) >droSim1.chr2R 9844261 52 + 19596830 ACAUCGAUGGUU----CCUUAAAAAGGAUCGAUUAUUGUGAUUUUUCGAAAACAUC .....((((..(----(((.....))))((((..(.......)..))))...)))) ( -8.10, z-score = 0.03, R) >droSec1.super_1 8599197 51 + 14215200 ACAACGAUGAUU----CCUUGAAAAACAUCGAUUAUUGUGAUUUU-CGAACACAGC ....(((((...----..........)))))....(((((.....-....))))). ( -5.92, z-score = -0.02, R) >droYak2.chr4 884476 56 + 1374474 ACAUCGAUGUUAUGAACUUCCAUAAACAUCAAUGUUAGCGAUGAUUGAAAAACAUU .(((((((((((((......))).))))))...(....)))))............. ( -10.40, z-score = -1.86, R) >droAna3.scaffold_3805 8894 52 - 10702 ACAUCGAUGUU---GGCUUGC-CAAACAUCGAUGUCAACGAUGUUUUCAAAACAUC (((((((((((---((....)-).)))))))))))....((((((.....)))))) ( -20.60, z-score = -5.06, R) >consensus ACAUCGAUGAU_____CCUUCAAAAACAUCGAUGAUUGCGAUUUUUCGAAAACAUC .((((((((.................))))))))...................... ( -4.37 = -5.37 + 1.00)

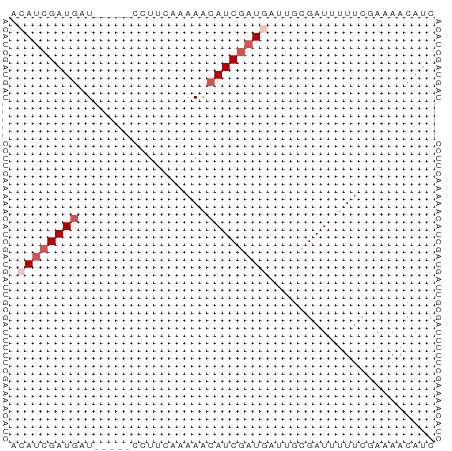
| Location | 11,105,810 – 11,105,861 |
|---|---|
| Length | 51 |
| Sequences | 5 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 67.84 |
| Shannon entropy | 0.56634 |
| G+C content | 0.34412 |
| Mean single sequence MFE | -12.56 |
| Consensus MFE | -5.25 |
| Energy contribution | -5.89 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
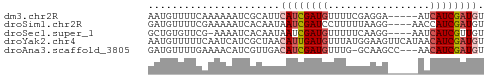
>dm3.chr2R 11105810 51 - 21146708 AAUGUUUUCAAAAAAUCGCAUUCAUCGAUGUUUUCGAGGA-----AUCAUCGAUGU ......................((((((((.(((....))-----).)))))))). ( -11.60, z-score = -1.38, R) >droSim1.chr2R 9844261 52 - 19596830 GAUGUUUUCGAAAAAUCACAAUAAUCGAUCCUUUUUAAGG----AACCAUCGAUGU ((((...((((.............))))((((.....)))----)..))))..... ( -7.72, z-score = -0.39, R) >droSec1.super_1 8599197 51 - 14215200 GCUGUGUUCG-AAAAUCACAAUAAUCGAUGUUUUUCAAGG----AAUCAUCGUUGU ..((((....-.....))))((((.(((((..(((....)----)).))))))))) ( -7.70, z-score = -0.20, R) >droYak2.chr4 884476 56 - 1374474 AAUGUUUUUCAAUCAUCGCUAACAUUGAUGUUUAUGGAAGUUCAUAACAUCGAUGU .....................(((((((((((.((((....))))))))))))))) ( -13.50, z-score = -2.51, R) >droAna3.scaffold_3805 8894 52 + 10702 GAUGUUUUGAAAACAUCGUUGACAUCGAUGUUUG-GCAAGCC---AACAUCGAUGU ((((((.....))))))....(((((((((((.(-(....))---))))))))))) ( -22.30, z-score = -4.84, R) >consensus GAUGUUUUCAAAAAAUCGCAAUCAUCGAUGUUUUUGAAGG_____AACAUCGAUGU ......................((((((((.................)))))))). ( -5.25 = -5.89 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:29 2011