| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,100,341 – 11,100,436 |
| Length | 95 |
| Max. P | 0.650000 |

| Location | 11,100,341 – 11,100,436 |
|---|---|
| Length | 95 |
| Sequences | 14 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 61.54 |
| Shannon entropy | 0.79711 |
| G+C content | 0.56617 |
| Mean single sequence MFE | -33.29 |
| Consensus MFE | -12.56 |
| Energy contribution | -12.83 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
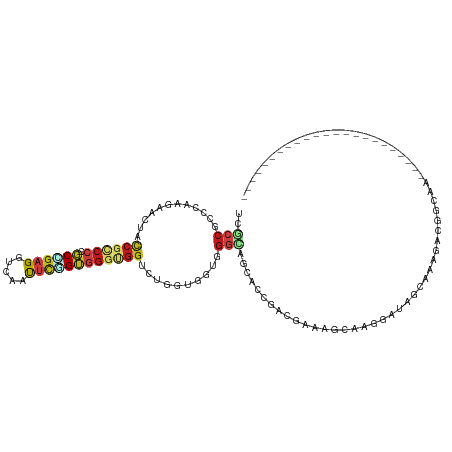
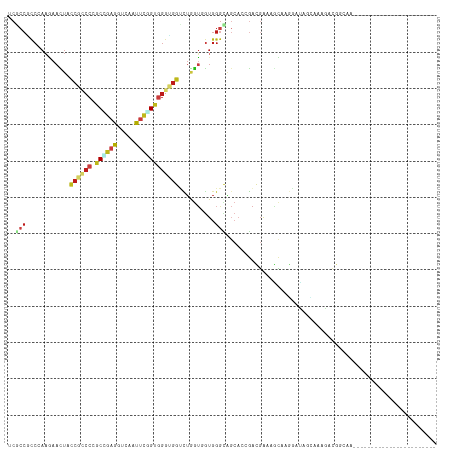
>dm3.chr2R 11100341 95 - 21146708 UCGCCGCCGAAGAACUACCGCCCCGCCGAGGUUAAUUCGGUGGGCGGUCUGGUGGUGGGCAGCACCGACGAAAGCAAGGAUAGCAAAGACGGCAA----------------------- ..((((((.(...((((((((((.((((((.....)))))))))))))..)))..).))).((.((..(....)...))...))......)))..----------------------- ( -41.10, z-score = -2.04, R) >droSim1.chr2R 9839306 95 - 19596830 UCGCCGCCAAAGAACUACCGCCCCGCCGAGGUUAAUUCGGUGGGCGGUCUGGUGGUGGGCAGCACCGACGAAAGCAAGGAUAGCAAAGACGGCAA----------------------- ..((((((.....((((((((((.((((((.....)))))))))))))..)))....))).((.((..(....)...))...))......)))..----------------------- ( -40.20, z-score = -2.05, R) >droSec1.super_1 8594275 95 - 14215200 UCGCCGCCCAAGAACUACCGCCCCGCCGAGGUUAAUUCGGUGGGCGGUCUGGUGGUGGGCAGCACCGACGAAAGCAAGGAUAGCAAAGACGGCAA----------------------- ..((((((((...((((((((((.((((((.....)))))))))))))..)))..))))).((.((..(....)...))...))......)))..----------------------- ( -45.50, z-score = -3.30, R) >droYak2.chr2R 11022451 95 - 21139217 UCGCCGCCGAAGAACUACCGCCCCGCCGAGGUUAAUUCAGUGGGCGGUCUGGUGGUGGGCAGCACGGACGAAAGCAAGGACAGCAAAGACGGCAA----------------------- .....((((.....(((((((((((((.(.(......)..).)))))...))))))))((.(..(...(....)...)..).)).....))))..----------------------- ( -33.90, z-score = -0.22, R) >droEre2.scaffold_4845 7822198 95 + 22589142 UCGCCACCGAAGAACUACCGCCCCGCCGAGGUUAAUUCAGUGGGUGGCCUGGUGGUGGGCAGCACCGACGAAAGCAAGGAUAGCAAAGACGGCAA----------------------- ..(((.........(((((((((((((.(.(......)..).)))))...))))))))...((.((..(....)...))...))......)))..----------------------- ( -30.80, z-score = 0.14, R) >droAna3.scaffold_13266 17635804 92 + 19884421 UCUCCUCCGAAAAAUUACCGCCCCGCUGAGGUCAACUCGGUGGGUGGGCUGGUGGUGGGAAGCUCGGAUGACAAUAAGGAUAAGGACAGCAA-------------------------- ..((.(((((....((((((((((((((((.....))))))))(....).)))))))).....))))).)).....................-------------------------- ( -30.50, z-score = -0.43, R) >dp4.chr3 14390574 95 + 19779522 UCUCCUCCUAAAAACUAUCGGCCAGCUGAGGUCAACUCGGUGGGAGGUCUAGUUGUGGGCAGCACUGACGAAAGUAAGGAUUCUAAAGACGUUAA----------------------- .(((((((...........))...((((((.....)))))))))))((((.((((....)))).((.((....)).))........)))).....----------------------- ( -24.90, z-score = 0.06, R) >droPer1.super_2 3441929 95 + 9036312 UCUCCUCCUAAAAACUAUCGGCCAGCUGAGGUCAACUCGGUGGGAGGUCUAGUUGUGGGCAGCACUGACGAAAGUAAGGAUUCUAAAGACGUUAA----------------------- .(((((((...........))...((((((.....)))))))))))((((.((((....)))).((.((....)).))........)))).....----------------------- ( -24.90, z-score = 0.06, R) >droWil1.scaffold_180699 2537996 101 - 2593675 CCGCCACCCAAAAACUAUCGCCCUGCCGAGGUGAAUUCAGUUGGUGGUCUUGUUGUGGGCAGCAACGAUGAUGCUAAAGAUGGCAAGGACAGCAAUAACAA----------------- ..((((((....((((.((((((....).)))))....))))))))))(((((..(.((((.((....)).))))....)..)))))..............----------------- ( -25.80, z-score = 0.65, R) >droVir3.scaffold_12875 9991183 101 - 20611582 GCGCCGCCCAAAAACUUCCGUCCUGCUGAGGUCAACUCUGUGGGUGGUCUGGUUGUGGGUAGCAACGAUGACAGUAGUGGUAAAGAUGGUAAGGAUUCUAA----------------- ..((..((((..((((.(((.((..(.(((.....))).)..)))))...)))).))))..))....((.((....)).))....................----------------- ( -28.50, z-score = 0.13, R) >droMoj3.scaffold_6496 9831807 101 + 26866924 GCUCCGCCCAAGAACUUCCGUCCUGCAGAGGUGAAUUCAGUGGGUGGUCUGGUAGUGGGCAGCAACGAUGACAGCGCGAGCAAAGAUGGCAAGGAUGCAAA----------------- ((((((((((...(((.(((.((..(.(((.....))).)..)))))...)))..))))).............((....))...........))).))...----------------- ( -32.90, z-score = -0.12, R) >droGri2.scaffold_15245 3175151 101 - 18325388 GCGCCACCCAAGAAUUUCCGUCCGGCAGAGGUCAAUUCUGUGGGUGGUUUGGUGGUGGGUAGCAACGAUGACAGCAGCGCCAAGAAUGGCAAAGAUGCCAA----------------- ..((((((((((((((..(.((.....)).)..)))))).))))))))((((((.((.((..........))..)).))))))...(((((....))))).----------------- ( -36.60, z-score = -1.24, R) >anoGam1.chr2R 62142791 72 + 62725911 ---------------------CCGCCGCAGAGC--CCUCGGCCGUUG-UCGAUCUU--ACG--AAAGAUGAU-UCGGGCAAACCGACGGCCGAUUCGCGAG----------------- ---------------------....(((.(((.--..(((((((..(-((.(((((--...--.))))))))-((((.....)))))))))))))))))..----------------- ( -26.70, z-score = -1.32, R) >triCas2.ChLG5 7919464 113 + 18847211 UCGACGUCUACCAAUUUUCGCCCGACGGAGAGCAACGUCGAGGGCGGCUUAAGCAUGGGUG--CAAAGCCGU-CGAAGAGUAUCGAU--CUGACUGGCGAAGAUGGUCGUGCGUUACC ..((((((.((((.(((((((((((((........)))))((((((((((..(((....))--).)))))))-)..(((.......)--))..)))))))))))))).).)))))... ( -43.70, z-score = -1.90, R) >consensus UCGCCGCCCAAGAACUACCGCCCCGCCGAGGUCAAUUCGGUGGGUGGUCUGGUGGUGGGCAGCACCGACGAAAGCAAGGAUAGCAAAGACGGCAA_______________________ ......((.........((((((.((((((.....)))))))))))).........))............................................................ (-12.56 = -12.83 + 0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:27 2011