| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,087,842 – 11,087,944 |
| Length | 102 |
| Max. P | 0.932319 |

| Location | 11,087,842 – 11,087,944 |
|---|---|
| Length | 102 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.46 |
| Shannon entropy | 0.63989 |
| G+C content | 0.40475 |
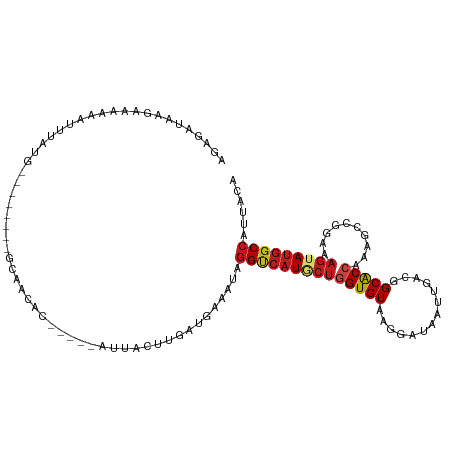
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -13.94 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11087842 102 + 21146708 AGAGAUAAGAUGGAAUUUAAG--------GCAACGC-----AUUACUUGAUGAAAUAGGACAUACUGGUGUACGGAUAAUUGACGGCACCAAAGCCAGAAAGUAUGGCCAUUACA ........(((((..(((..(--------((....(-----(((....)))).............((((((.((.........))))))))..)))..)))......)))))... ( -19.70, z-score = -1.00, R) >droSim1.chr2R 9826719 102 + 19596830 AGAGAUAAAAACGAAUUUAUG--------GCAACAC-----AUUACUUAAUGAAAUAGGCCAUACUGGUGUACGGAUAAUUGACGGCACCAAAGCCGGAAAGUAUGGCCAUUACA ...................((--------....))(-----(((....)))).....((((((((((.....)..........((((......))))...)))))))))...... ( -24.40, z-score = -2.38, R) >droSec1.super_1 8581757 102 + 14215200 AGAGAUAAGAAGGAAUUUAUG--------GCAACAC-----AUUACUUAAUGAAAUAGGCCAUACUGGUGUACGGAUAAUUAACGGCACCAAAGCCGGAAAGUAUGGCCAUUACA ...................((--------....))(-----(((....)))).....((((((((((.....)..........((((......))))...)))))))))...... ( -24.40, z-score = -2.33, R) >droYak2.chr2R 11009794 102 + 21139217 AGAGAUAAGAAUGAAUUUAUG--------GCAACCC-----AUUACUUGAUGAAAUAGGCCAUACUGGUGUACGGAUAAUUGACGGCGCCAAAGCCCGAAAGUAUGGCCAUAACA .....((((((((........--------......)-----))).))))........((((((((((((((.((.........)))))))..........)))))))))...... ( -23.04, z-score = -1.13, R) >droEre2.scaffold_4845 7809453 102 - 22589142 AGAGAUAAGAAGAAAUUUAUG--------GCAACUC-----AUUACUUGAUGAAAUAGGUCAUACUGGUGUACGGAUAAUUGACGGCACCAAAGCCGGAAAGUAUGGCCAUUACA ........(((....)))..(--------....)((-----(((....)))))....((((((((((.....)..........((((......))))...)))))))))...... ( -22.30, z-score = -1.23, R) >droAna3.scaffold_13266 17623348 104 - 19884421 UAAGAUAAGAACAAAUUUGUA--------GCCACUCCA---AUUACUUGAUAAAAUAGGUCAUGCUGGUGUAAGGAUAGUUCACAGCACCAAAGCCCGAAAGGAUGGCCAUUACG ..................(((--------(......((---(....)))........((((((((((((((...((....))...)))))..)))((....)))))))).)))). ( -22.70, z-score = -1.15, R) >dp4.chr3 14377717 104 - 19779522 AAUGAUAAGAAACGGUUUAGA--------GCAGUUAAC---AUUACUUGAUAAAGUAGGUCAUGCUGGUGUAAGGAUAGUUCACGGCACCAAAUCCAGACAGUAUGGCCAUUACA ..........(((.((.....--------)).)))...---..(((((....)))))((((((((((((((...((....))...)))))..........)))))))))...... ( -23.60, z-score = -1.08, R) >droPer1.super_2 3429110 104 - 9036312 AAUGAUAAGAAACGUUUUAGA--------GCAGUUAAC---AUUACUUGAUAAAGUAGGUCAUGCUGGUGUAAGGAUAGUUCACGGCACCAAAUCCAGACAGUAUGGCCAUUACA ..((.(((((....)))))..--------.))......---..(((((....)))))((((((((((((((...((....))...)))))..........)))))))))...... ( -23.20, z-score = -1.12, R) >droWil1.scaffold_180699 2525976 88 + 2593675 -----------------------AAA---AUUCUGAUGAUGUU-ACUUAAUGAAAUAGGUCAUGCUGGUGUAAGGAUAAUUUACAGCACCAAAGCCCGAAAGUAUAGCCAUUACG -----------------------...---....(((((..(((-(............(((..((.((.((((((.....)))))).)).))..)))(....)..))))))))).. ( -14.40, z-score = 0.01, R) >droVir3.scaffold_12875 9982384 109 + 20611582 GCUGAUAAGAACACAGCUGUGUACGG---GCGAUGGUG--ACU-ACUUGAUAAAAUAGGUCAUGCUGGUGUAUGGAUAGUUGACAGCACCAAAGCCGGAGAGUAUGGCCAUGACA .(((......((((....)))).(((---(....(...--.).-.))))......)))((((((.((((((.((.........))))))))..((((.......)))))))))). ( -26.20, z-score = 0.59, R) >droMoj3.scaffold_6496 9822902 108 - 26866924 UCAGAUAAGUCUACAGUUGUGUGUGG---GCU-CGAUG--GGU-ACUUGAUAAAAUAGGUCAUGCUGGUGUAUGGAUAAUUGACAGCACCAAAGCCGGAGAGAAUGGCCAUGACA (((....((((((((......)))))---)))-...))--)..-..............((((((.((((((.((.........))))))))..((((.......)))))))))). ( -28.50, z-score = -0.34, R) >droGri2.scaffold_15245 3166427 115 + 18325388 AGUGAUAAGAAAUCAUUUAUGUGUUGUGUGUUUUAUGGACGCUUACUUAAUGAAAUAGGUCAUGCUAGUGUAUGGAUAAUUGACAGCACCGAAGCCGGAUAGUAUGGCCAUGACU (((.((......(((((...(((....(((((.....))))).)))..)))))....((((((((((.(((.((.........)))))(((....))).)))))))))))).))) ( -27.90, z-score = -0.86, R) >triCas2.chrUn_23 31933 97 - 150934 -----------AUUGUUUUCGACU-----GAAUUAAAAU--AGAACCUAAUGAAGUAAGCCAUAGAUGUGUAAGGGUAGUUAACGGCACCAAACCCCGAAAGGAGGGCCAUUACA -----------.((((.....(((-----..((((....--......))))..))).....))))...(((((.(((.((.....))......((((....)).))))).))))) ( -15.50, z-score = 1.02, R) >consensus AGAGAUAAGAAAAAAUUUAUG________GCAACAC_____AUUACUUGAUGAAAUAGGUCAUGCUGGUGUAAGGAUAAUUGACGGCACCAAAGCCGGAAAGUAUGGCCAUUACA .........................................................((((((((((((((..............)))))..........)))))))))...... (-13.94 = -14.38 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:26 2011