| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,057,478 – 11,057,588 |
| Length | 110 |
| Max. P | 0.582165 |

| Location | 11,057,478 – 11,057,588 |
|---|---|
| Length | 110 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Shannon entropy | 0.39921 |
| G+C content | 0.48131 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -15.81 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
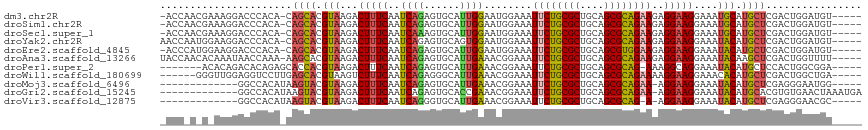
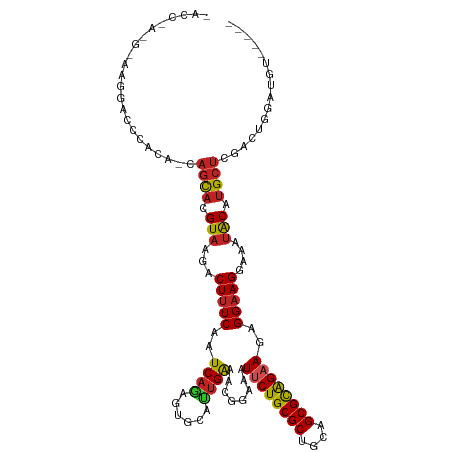
>dm3.chr2R 11057478 110 + 21146708 -----ACAUCCAGUCGAGCAUGCAUUUCCUUCCUCUUCUGCGCUGCAGCGCAGAAUUUCCAUUCCAAUGCACUCUGAUUGAAAGUCUUACGUGCUG-UGUGGGUCCUUUCGUUGGU- -----....(((((((((..((((((.........((((((((....))))))))..........))))))))).))))(((((.((((((.....-))))))..)))))...)).- ( -31.41, z-score = -1.54, R) >droSim1.chr2R 9795973 110 + 19596830 -----ACAUCCAGUCGAGCAUGCAUUUCCUUCCUCUUCUGCGCUGCAGCGCAGAAUUUCCAUUCCAAUGCACUCUGAUUGAAAGUCUUACGUGCUG-UGUGGGUCCUUUCGUUGGU- -----....(((((((((..((((((.........((((((((....))))))))..........))))))))).))))(((((.((((((.....-))))))..)))))...)).- ( -31.41, z-score = -1.54, R) >droSec1.super_1 8551560 110 + 14215200 -----ACAUCCAGUCGAGCAUGCAUUUCCUUCCUCUUCUGCGCUGCAGCGCAGAAUUUCCAUUCCAAUGCACUUUGAUUGAAAGUCUUACGUGCUG-UGUGGGUCCUUUCGUUGGU- -----....(((((((((..((((((.........((((((((....))))))))..........)))))).)))))))(((((.((((((.....-))))))..)))))...)).- ( -31.21, z-score = -1.56, R) >droYak2.chr2R 10978677 111 + 21139217 -----ACAUCCAGUCGAGCAUGUAUUUCCUUCCUCUUCUGCGCUGCAGCGCAGAAUUUCCAUUCCACUGCACUCUGAUUGAAAGUCUUACGUGCUG-UGUGGGUCCUUCCAUUGGUU -----....(((((.(((.................((((((((....)))))))).......(((((.((((...(((.....)))....))))..-.)))))...))).))))).. ( -31.10, z-score = -1.95, R) >droEre2.scaffold_4845 7772737 110 - 22589142 -----ACAUCCAGUCGAGCAUGUAUUUCCUUCCUCUUCCACGCUGCAGCGCAGAAUUUCCAUUCCAAUGCACUCUGAUUGAAAGUCUUACGUGCUG-UGUGGGUCCUUCCAUGGGU- -----..((((.....((((((((...(.(((........(((....)))((((.....(((....)))...))))...))).)...)))))))).-.(((((....)))))))))- ( -23.50, z-score = 0.68, R) >droAna3.scaffold_13266 17592982 111 - 19884421 -----AAAACCAGUCGAGCUUGUAUUUCCUUCCUCUUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACUCUGAUUGAAAGUCUUACGUGCUU-UUUGGUUAUUUGUGUUGGUA -----..((((((..((((.((((...........((((((((....))))))))......(((((((........)))))))....)))).))))-.))))))............. ( -29.60, z-score = -2.29, R) >droPer1.super_2 3400289 104 - 9036312 -----UCCGCCAGUGGAGCAUGUAUUUCCUGCCUUU-CUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACUCUGAUUGAAAGUCUUACGUGGUGCUCUGUGUCUGUGU------- -----..(((..(..((((((.............((-((((((....))))))))......(((((((........)))))))..........))))))..)....))).------- ( -29.70, z-score = -0.97, R) >droWil1.scaffold_180699 2482929 106 + 2593675 -----UCAGCCAGUCGAGCAUGUGUUUCCUUCCUUUUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCCCUCUGAUUGAAAGACUUACGUGCUCAAGGACCUCCAACCC------ -----..........(((((((((..((.......((((((((....))))))))......(((((((........)))))))))..)))))))))..((....)).....------ ( -34.10, z-score = -3.75, R) >droMoj3.scaffold_6496 9771941 98 - 26866924 -----CCAUUCCCUCGAGCAUGUAUUUCCUUCCU-UUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACUCUGAUUGAAAGUCUUACGUACUUAUGUGGCC------------- -----((((......(((.(((((..........-((((((((....))))))))......(((((((........)))))))....))))).)))..))))..------------- ( -23.50, z-score = -2.17, R) >droGri2.scaffold_15245 3129752 103 + 18325388 UCAUUUAGUUCACACGUGCAUGUAUUUCCUUCCU-UUCUGCGCUGCAGCGCAGAAUUUCCGUUUCGGUGCACUCUGAUUGAAAGUCUUACGUACUUAUGUGGCC------------- ............((((((.(((((..........-((((((((....))))))))((((....((((......))))..))))....)))))...))))))...------------- ( -24.70, z-score = -1.09, R) >droVir3.scaffold_12875 9937795 97 + 20611582 -----GCGUUCCCUCGAGCAUGUAUUUCCUUCCU-U-CUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACCCUGAUUGAAAGUCUUACGUACUUAUGUGGCC------------- -----((((((....))))(((((.........(-(-((((((....))))))))......(((((((........)))))))....))))).........)).------------- ( -23.50, z-score = -1.65, R) >consensus _____ACAUCCAGUCGAGCAUGUAUUUCCUUCCUCUUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACUCUGAUUGAAAGUCUUACGUGCUG_UGUGGGUCCUU_C_U_GGU_ ...............(((..((((((.........((((((((....))))))))..........)))))))))........................................... (-15.81 = -16.02 + 0.21)

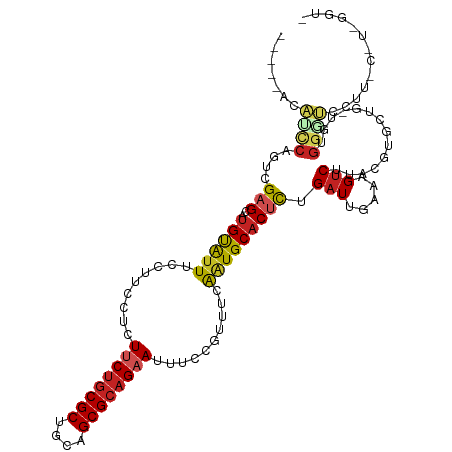
| Location | 11,057,478 – 11,057,588 |
|---|---|
| Length | 110 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.76 |
| Shannon entropy | 0.39921 |
| G+C content | 0.48131 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -20.93 |
| Energy contribution | -20.79 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 11057478 110 - 21146708 -ACCAACGAAAGGACCCACA-CAGCACGUAAGACUUUCAAUCAGAGUGCAUUGGAAUGGAAAUUCUGCGCUGCAGCGCAGAAGAGGAAGGAAAUGCAUGCUCGACUGGAUGU----- -.(((..(((((.....((.-......))....))))).....(((((((((..........((((((((....)))))))).........)))))..))))...)))....----- ( -29.41, z-score = -0.64, R) >droSim1.chr2R 9795973 110 - 19596830 -ACCAACGAAAGGACCCACA-CAGCACGUAAGACUUUCAAUCAGAGUGCAUUGGAAUGGAAAUUCUGCGCUGCAGCGCAGAAGAGGAAGGAAAUGCAUGCUCGACUGGAUGU----- -.(((..(((((.....((.-......))....))))).....(((((((((..........((((((((....)))))))).........)))))..))))...)))....----- ( -29.41, z-score = -0.64, R) >droSec1.super_1 8551560 110 - 14215200 -ACCAACGAAAGGACCCACA-CAGCACGUAAGACUUUCAAUCAAAGUGCAUUGGAAUGGAAAUUCUGCGCUGCAGCGCAGAAGAGGAAGGAAAUGCAUGCUCGACUGGAUGU----- -.....(....)...(((..-((((.(....).............(((((((..........((((((((....)))))))).........)))))))))).)..)))....----- ( -28.31, z-score = -0.49, R) >droYak2.chr2R 10978677 111 - 21139217 AACCAAUGGAAGGACCCACA-CAGCACGUAAGACUUUCAAUCAGAGUGCAGUGGAAUGGAAAUUCUGCGCUGCAGCGCAGAAGAGGAAGGAAAUACAUGCUCGACUGGAUGU----- ..(((.(((......)))((-(.((((..................)))).)))...)))...((((((((....))))))))(((.(.(......).).)))..........----- ( -29.27, z-score = -0.74, R) >droEre2.scaffold_4845 7772737 110 + 22589142 -ACCCAUGGAAGGACCCACA-CAGCACGUAAGACUUUCAAUCAGAGUGCAUUGGAAUGGAAAUUCUGCGCUGCAGCGUGGAAGAGGAAGGAAAUACAUGCUCGACUGGAUGU----- -..(((.........((((.-(.((.(....)............((((((...((((....)))))))))))).).))))..(((.(.(......).).)))...)))....----- ( -24.80, z-score = 0.92, R) >droAna3.scaffold_13266 17592982 111 + 19884421 UACCAACACAAAUAACCAAA-AAGCACGUAAGACUUUCAAUCAGAGUGCAUUGAAACGGAAAUUCUGCGCUGCAGCGCAGAAGAGGAAGGAAAUACAAGCUCGACUGGUUUU----- .............(((((..-.(((..(((...(((((..((...((........)).))..((((((((....))))))))..)))))....)))..)))....)))))..----- ( -26.30, z-score = -1.76, R) >droPer1.super_2 3400289 104 + 9036312 -------ACACAGACACAGAGCACCACGUAAGACUUUCAAUCAGAGUGCAUUGAAACGGAAAUUCUGCGCUGCAGCGCAG-AAAGGCAGGAAAUACAUGCUCCACUGGCGGA----- -------...(((...(((.((((..(....)....((.....)))))).)))....(((..((((((((....))))))-))..((((......).)))))).))).....----- ( -28.20, z-score = -1.16, R) >droWil1.scaffold_180699 2482929 106 - 2593675 ------GGGUUGGAGGUCCUUGAGCACGUAAGUCUUUCAAUCAGAGGGCAUUGAAACGGAAAUUCUGCGCUGCAGCGCAGAAAAGGAAGGAAACACAUGCUCGACUGGCUGA----- ------.(((..(......(((((((.((...((((((..((((......))))........((((((((....))))))))...))))))...)).))))))))..)))..----- ( -34.30, z-score = -1.34, R) >droMoj3.scaffold_6496 9771941 98 + 26866924 -------------GGCCACAUAAGUACGUAAGACUUUCAAUCAGAGUGCAUUGAAACGGAAAUUCUGCGCUGCAGCGCAGAA-AGGAAGGAAAUACAUGCUCGAGGGAAUGG----- -------------..((.....((((.(((...(((((..((...((........)).))..((((((((....))))))))-.)))))....))).))))....)).....----- ( -24.80, z-score = -1.27, R) >droGri2.scaffold_15245 3129752 103 - 18325388 -------------GGCCACAUAAGUACGUAAGACUUUCAAUCAGAGUGCACCGAAACGGAAAUUCUGCGCUGCAGCGCAGAA-AGGAAGGAAAUACAUGCACGUGUGAACUAAAUGA -------------(..(((((((((.(....))))).........((((((((...)))...((((((((....))))))))-..............))))))))))..)....... ( -30.30, z-score = -2.65, R) >droVir3.scaffold_12875 9937795 97 - 20611582 -------------GGCCACAUAAGUACGUAAGACUUUCAAUCAGGGUGCAUUGAAACGGAAAUUCUGCGCUGCAGCGCAG-A-AGGAAGGAAAUACAUGCUCGAGGGAACGC----- -------------..((.....((((.(((...(((((..((((......))))........((((((((....))))))-)-))))))....))).))))....)).....----- ( -25.50, z-score = -1.15, R) >consensus _ACC_A_G_AAGGACCCACA_CAGCACGUAAGACUUUCAAUCAGAGUGCAUUGAAACGGAAAUUCUGCGCUGCAGCGCAGAAGAGGAAGGAAAUACAUGCUCGACUGGAUGU_____ ......................((((.(((...(((((..((((......))))........((((((((....))))))))..)))))....))).))))................ (-20.93 = -20.79 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:25 2011