| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,051,855 – 11,051,947 |
| Length | 92 |
| Max. P | 0.595438 |

| Location | 11,051,855 – 11,051,947 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 61.38 |
| Shannon entropy | 0.77676 |
| G+C content | 0.34052 |
| Mean single sequence MFE | -13.09 |
| Consensus MFE | -4.21 |
| Energy contribution | -4.18 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595438 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
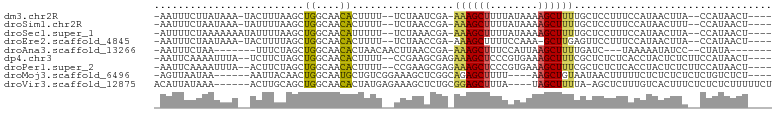
>dm3.chr2R 11051855 92 + 21146708 -AAUUUCUUAUAAA-UACUUUAAGCUGGCAACACUUUU--UCUAAUCGA-AAAGCUUUUAUAAAAGCUUUUGCUCCUUUCCAUAACUUA--CCAUAACU---- -.............-.......(((((....)).....--........(-((((((((....))))))))))))...............--........---- ( -10.90, z-score = -1.62, R) >droSim1.chr2R 9790189 92 + 19596830 -AAUUUCUAAUAAA-UAUUUUAAGCUGGCAACACUUUU--UCUAACCGA-AAAGCUUUUAUAAAAGCUUUUGCUCCUUUCCAUAACUUU--CCAUAACU---- -.............-.......(((((....)).....--........(-((((((((....))))))))))))...............--........---- ( -10.90, z-score = -1.49, R) >droSec1.super_1 8545810 93 + 14215200 -AUUUUCUAAAAAAAUAUUUUAAGCUGGCAACAUUUUU--UCUAAACGA-AAAGCUUUUAUAAAAGCUUUUGCUCCUUUCCAUAACUUA--CCAUAACU---- -.....................(((((....)).....--........(-((((((((....))))))))))))...............--........---- ( -11.00, z-score = -1.21, R) >droEre2.scaffold_4845 7766970 91 - 22589142 -AAUUUCUAAUAAA-UACUUUUAGCUGGCAACACUUUU--UCUAACCGA-AAAGCUUUUCCAAA-GCUUGAGUUCCUUUCCAUAACUUA--CCAUAACU---- -.............-.((((..((((((.((..(((((--(......))-))))..)).))..)-))).))))................--........---- ( -9.30, z-score = -0.85, R) >droAna3.scaffold_13266 17587281 82 - 19884421 -AAUUUCUAA-------UUUCUAGCUGGCAACACUAACAACUUAACCGA-AAAGCUUUCCAUUAAGCUUUUGAUC---UAAAAAUAUCC--CUAUA------- -.........-------....(((.((....)))))............(-(((((((......))))))))....---...........--.....------- ( -10.80, z-score = -2.19, R) >dp4.chr3 14336628 94 - 19779522 -AAUUCAAAAUUUA--UCUUCUAGCUGGCAACACUUUU--CCGAAGCGAGAAAGCUCCCGUGAAAGCUUUCGCUCUCUCACCUACUCUCUUCCAUAACU---- -.............--.((((.((.((....))))...--..)))).((((.(((....((....))....))).))))....................---- ( -13.90, z-score = -0.59, R) >droPer1.super_2 3393878 94 - 9036312 -AAUUCAAAAUUUA--ACUUCUAGCUGGCAACACUUUU--CCGAAGCGAGAAAGCUCCCGUGAAAGCUUUCGCUCUCUCACCUACUCUCUUCCAUAACU---- -.............--.((((.((.((....))))...--..)))).((((.(((....((....))....))).))))....................---- ( -14.30, z-score = -0.78, R) >droMoj3.scaffold_6496 9761036 88 - 26866924 -AGUUAAUAA------AAUUACAACUGGCAAUGCUGUCGGAAAGCUCGGCAGAGCUUUU----AAGCUGUAAUAACUUUUUCUCUCUCUCUCUGUCUCU---- -.........------.((((((.((((((....))))))(((((((....))))))).----....))))))..........................---- ( -18.40, z-score = -1.40, R) >droVir3.scaffold_12875 9929831 92 + 20611582 ACAUUAUAAA------ACUUGCAGCUGGCAACACUAUGAGAAAGCUCUGCGGAGCUUUA----UAGCUUUUA-AGCUCUUUGUCACUUUCUCUCUCUUUUUCU ..........------......((.((....))))..(((((((......(((((((..----........)-))))))......)))))))........... ( -18.30, z-score = -0.55, R) >consensus _AAUUUCUAAUAAA__ACUUUUAGCUGGCAACACUUUU__UCUAACCGA_AAAGCUUUUAUAAAAGCUUUUGCUCCUUUCCAUAACUUA__CCAUAACU____ .........................((....)).................((((((........))))))................................. ( -4.21 = -4.18 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:23 2011