| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,048,972 – 11,049,053 |
| Length | 81 |
| Max. P | 0.986702 |

| Location | 11,048,972 – 11,049,053 |
|---|---|
| Length | 81 |
| Sequences | 11 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 71.07 |
| Shannon entropy | 0.61617 |
| G+C content | 0.37359 |
| Mean single sequence MFE | -15.31 |
| Consensus MFE | -8.41 |
| Energy contribution | -8.53 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
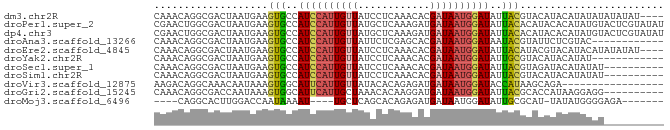
>dm3.chr2R 11048972 81 + 21146708 CAAACAGGCGACUAAUGAAGUGCCAUCCAUUGUUAUCCUCAAACACGAUAAUGGAUAUUACGUACAUACAUAUAUAUAUAU---- ...((.(((.(((.....))))))((((((((((............)))))))))).....))..................---- ( -14.90, z-score = -1.61, R) >droPer1.super_2 3390914 85 - 9036312 CGAACUGGCGACUAAUGAAGUGCCAUCCAUUGUUAUGCUCAAAGAUGAUAAUGGAUAUUACACAUACACAUAUGUACUCGUAUAU (((...(((.(((.....))))))((((((((((((........)))))))))))).....(((((....)))))..)))..... ( -20.70, z-score = -2.12, R) >dp4.chr3 14333647 85 - 19779522 CGAACUGGCGACUAAUGAAGUGCCAUCCAUUGUUAUGCUCAAAGAUGAUAAUGGAUAUUACACAUACACAUAUGUACUCGUAUAU (((...(((.(((.....))))))((((((((((((........)))))))))))).....(((((....)))))..)))..... ( -20.70, z-score = -2.12, R) >droAna3.scaffold_13266 17584439 73 - 19884421 CAAACAGGCGACUAAUGAAGUGCCAUCCAUUGUUAUUCUCGAGCACGAUAAUGGAUAAUACGUAUUCUCGUAC------------ ......(((.(((.....))))))(((((((((..(.(....).)..)))))))))..((((......)))).------------ ( -17.50, z-score = -1.91, R) >droEre2.scaffold_4845 7763999 81 - 22589142 CAAACAGGCGACUAAUGAAGUGCCAUCCAUUGUUAUCCUCAAACACGAUAAUGGAUAUUACAUACGUACAUACAUAUAUAU---- ...((.(((.(((.....))))))((((((((((............)))))))))).........))..............---- ( -14.90, z-score = -1.77, R) >droYak2.chr2R 10969697 73 + 21139217 CAAACAGGCGACUAAUGAAGUGCCAUCCAUUGUUAUCCUCAAACACGAUAAUGGAUAUUGCGUACAUACAUAU------------ ...((.(((.(((.....))))))((((((((((............)))))))))).....))..........------------ ( -14.90, z-score = -1.56, R) >droSec1.super_1 8542936 75 + 14215200 CAAACAGGCGACUAAUGAAGUGCCAUCCAUUGUUAUCCUCAAACACGAUAAUGGAUAUUACGUAGAUACAUAUAU---------- ...((.(((.(((.....))))))((((((((((............)))))))))).....))............---------- ( -14.90, z-score = -1.86, R) >droSim1.chr2R 9787344 75 + 19596830 CAAACAGGCGACUAAUGAAGUGCCAUCCAUUGUUAUCCUCAAACACGAUAAUGGAUAUUACGUACAUACAUAUAU---------- ...((.(((.(((.....))))))((((((((((............)))))))))).....))............---------- ( -14.90, z-score = -2.02, R) >droVir3.scaffold_12875 9926757 68 + 20611582 AAGACAGGCAAACAAUAAAGUGGCAUUCAUUGUUAUACACAGAGAUGAUAAUGGAUACCAUAAGCAGA----------------- ...................((((.((((((((((((........)))))))))))).)))).......----------------- ( -15.00, z-score = -3.63, R) >droGri2.scaffold_15245 3118776 75 + 18325388 CAAACAGGCGACCAAUAAAGUGGCAUUCAUUGCUAAACACAAGGAUGAUAAUGGAUAUUACGCACCAUAAGGAGG---------- .......(((.((.......(((((.....))))).......)).(((((.....)))))))).((....))...---------- ( -11.54, z-score = -0.68, R) >droMoj3.scaffold_6496 9757486 69 - 26866924 ----CAGGCACUUGGACCAAUAAAAU----UGCUCAGCACAGAGAUGAUAAUGGAUAUUGCGCAU-UAUAUGGGGAGA------- ----......(((...(((.......----..(((......)))...(((((((......).)))-))).))).))).------- ( -8.50, z-score = 1.26, R) >consensus CAAACAGGCGACUAAUGAAGUGCCAUCCAUUGUUAUCCUCAAACACGAUAAUGGAUAUUACGUACAUACAUAUAU__________ ...................(((..((((((((((............))))))))))..)))........................ ( -8.41 = -8.53 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:21 2011