| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,025,850 – 11,025,968 |
| Length | 118 |
| Max. P | 0.978552 |

| Location | 11,025,850 – 11,025,968 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 126 |
| Reading direction | reverse |
| Mean pairwise identity | 77.38 |
| Shannon entropy | 0.49141 |
| G+C content | 0.60137 |
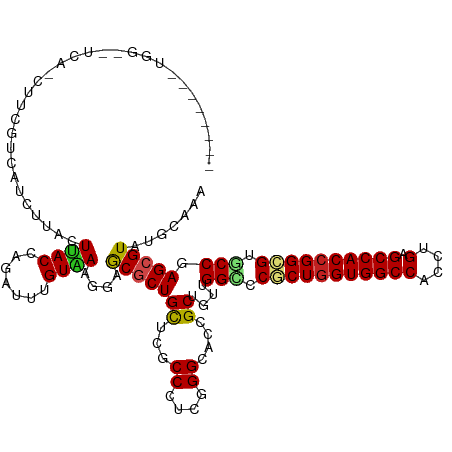
| Mean single sequence MFE | -45.50 |
| Consensus MFE | -31.77 |
| Energy contribution | -31.69 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
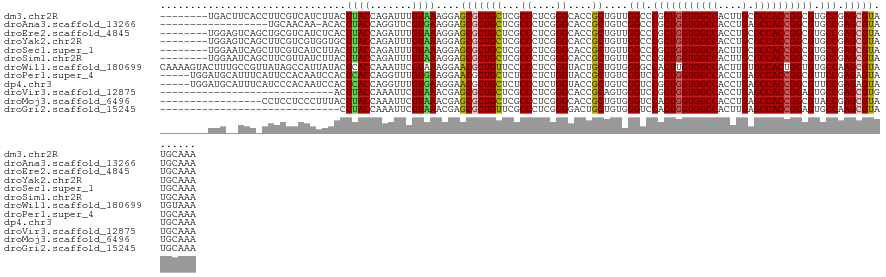
>dm3.chr2R 11025850 118 - 21146708 --------UGACUUCACCUUCGUCAUCUUACUUACCAGAUUUGUAAAGGAGCGCUGCUCGCCCUCGGGCACCGCUGUUGGCCCGCUGGUGGCCACUUGCGCCACCGGCGUGCCGAGCGUAUGCAAA --------........((((..(.((((........))))..)..))))((((.((((((....)))))).)))).(((((.(((((((((((....).)))))))))).)))))((....))... ( -47.70, z-score = -2.25, R) >droAna3.scaffold_13266 17562022 107 + 19884421 ------------------UGCAACAA-ACACUUACCAGGUUCGUGAAGGAGCGCUGCUCGCCCUCGGGCACCGCUGUCGGCCCGCUGGUGGCCACCUGAGCCACCGGCGUGCCGAGCGUAUGCAAA ------------------((((((..-.(((..((...))..)))....((((.((((((....)))))).)))).(((((.(((((((((((....).)))))))))).)))))..)).)))).. ( -50.00, z-score = -2.10, R) >droEre2.scaffold_4845 7740797 118 + 22589142 --------UGGAGUCAGCUGCGUCAUCUCACUUACCAGAUUUGUAAAGGAGCGCUGCUCGCCCUCGGGCACCGCUGUUGGCCCGCUGGUGGCCACCUGCGCCACCGGCGUGCCGAGCGUAUGCAAA --------.((.((.(((.(((....(((..((((.......))))..)))))).))).))))....(((.((((..((((.(((((((((((....).)))))))))).))))))))..)))... ( -49.90, z-score = -0.94, R) >droYak2.chr2R 10946312 118 - 21139217 --------UGGAGUCAGCUUCGUCGUGGUGCUUACCAGAUUUGUAAAGGAGCGCUGCUCGCCCUCGGGCACCGCUGUUGGCCCGCUGGUGGCCACCUGCGCCACCGGCGUGCCGAGCGUAUGCAAA --------.((((....))))(((.((((....)))))))(((((.(.(((((.((((((....)))))).)))).(((((.(((((((((((....).)))))))))).))))).).).))))). ( -52.90, z-score = -1.06, R) >droSec1.super_1 8520189 118 - 14215200 --------UGGAAUCAGCUUCGUCAUCUUACUUACCAGAUUUGUAAAGGAGCGCUGCUCGCCCUCGGGCACCGCUGUUGGCCCGCUGGUGGCCACUUGCGCCACCGGCGUGCCGAGCGUAUGCAAA --------........(((((..(((((........)))..))....)))))((((((((((....))).........(((.(((((((((((....).)))))))))).))))))))...))... ( -47.90, z-score = -1.48, R) >droSim1.chr2R 9764601 118 - 19596830 --------UGGAAUCAGCUUCGUUAUCUUACUUACCAGAUUUGUAAAGGAGCGCUGCUCGCCCUCGGGCACCGCUGUUGGCCCGCUGGUGGCCACUUGCGCCACCGGCGUGCCGAGCGUAUGCAAA --------........(((((...((((........)))).......)))))((((((((((....))).........(((.(((((((((((....).)))))))))).))))))))...))... ( -48.50, z-score = -1.73, R) >droWil1.scaffold_180699 2442252 126 - 2593675 CAAAAGUACUUUGCCGUUAUAGCCAUUAUACUCACCAAAUUCGUAAAGGAACGCUGUUCCCCCUCCGGUACUGCUGUGGGGCCACUUGUGGCCACUUGUGCCACUGGUGUGCCAAGCGUAUGUAAA .....((((..(((((............(((...........)))..(((((...))))).....)))))..(((....(((((((.((((((....).))))).)))).))).)))))))..... ( -34.60, z-score = 0.13, R) >droPer1.super_4 2778241 121 - 7162766 -----UGGAUGCAUUUCAUUCCACAAUCCACUCACCAGGUUUGUGAAGGAACGCUGCUCUCCCUCUGGUACCGCUGUCGGUCCGCUGGUGGCCACCUGAGCCACCGGCGUUCCGAGAGUAUGCAAA -----((.((((.((((....(((((.((........)).)))))..(((((..............((.((((....))))))((((((((((....).)))))))))))))))))))))).)).. ( -48.30, z-score = -2.95, R) >dp4.chr3 8892861 121 - 19779522 -----UGGAUGCAUUUCAUCCCACAAUCCACUCACCAGGUUUGUGAAGGAACGCUGCUCUCCCUCUGGUACCGCUGUCGGUCCGCUGGUGGCCACCUGAGCCACCGGCGUUCCGAGAGUAUGCAAA -----.(((((.....)))))(((((.((........)).))))).......(((((((((.....((.((((....))))))((((((((((....).))))))))).....))))))).))... ( -49.50, z-score = -3.22, R) >droVir3.scaffold_12875 9892550 97 - 20611582 -----------------------------ACUUACCAAAUUCGUAAACGAGCGCUGCUCGCCCUCGGGCACCGCAGUGGGUCCGCUGGUGGCCACCUGAGCCACCGGAGUGCCGAGCGUGUGCAAA -----------------------------...........(((....)))((((((((((((....))).........((..(.(((((((((....).)))))))).)..))))))).))))... ( -42.30, z-score = -1.63, R) >droMoj3.scaffold_6496 9724684 109 + 26866924 -----------------CCUCCUCCCUUUACUUACCAAAUUCGUAAACGAGCGCUGCUCGCCCUCGGGCACCGCUGUGGGUCCACUGGUGGCCACCUGAGCCACCGGCGUACCGAGCGUAUGCAAA -----------------.........................(((.(((((((.((((((....)))))).))))...(((.(.(((((((((....).)))))))).).)))...))).)))... ( -38.90, z-score = -1.73, R) >droGri2.scaffold_15245 3093636 96 - 18325388 ------------------------------CUUACCAAAUUCGUAAACGAGCGCUGUUCGCCCUCGGGGACUGCUGUGGGUCCACUGGUGGCCACUUGAGCCACCGGAGUGCCAAGCGUAUGCAAA ------------------------------............(((.(((.(((((....((((.(((......))).))))...(((((((((....).)))))))))))))....))).)))... ( -35.50, z-score = -1.05, R) >consensus ________UGG__UCA_CUUCGUCAUCUUACUUACCAGAUUUGUAAAGGAGCGCUGCUCGCCCUCGGGCACCGCUGUUGGCCCGCUGGUGGCCACCUGAGCCACCGGCGUGCCGAGCGUAUGCAAA ...............................((((.......))))....(((((((...((....))....))....(((.(((((((((((....).)))))))))).))).)))))....... (-31.77 = -31.69 + -0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:19 2011