
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 11,024,232 – 11,024,394 |
| Length | 162 |
| Max. P | 0.935385 |

| Location | 11,024,232 – 11,024,341 |
|---|---|
| Length | 109 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.46 |
| Shannon entropy | 0.61006 |
| G+C content | 0.63218 |
| Mean single sequence MFE | -48.23 |
| Consensus MFE | -18.35 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
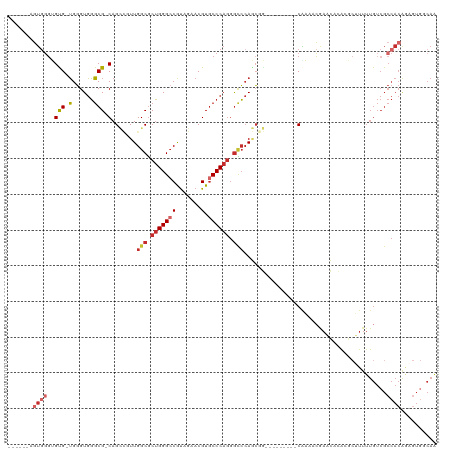
>dm3.chr2R 11024232 109 + 21146708 GCCACCAGUGGGUGUG-UGGGUGGGCG-CAGCCGUUGGCGCUGGGCCGUAGAACGUGCCCAGGGCCCAGCGG---------CAAAUCGAACACACGCUUUUUGUCGCCACAGAGUGGCAG ((((((.(((((((((-((..(((...-..(((((((((.((((((((.....)).)))))).).)))))))---------)...)))..))))))).........)))).).))))).. ( -58.80, z-score = -3.23, R) >droSim1.chr2R 9763007 109 + 19596830 GCCAACAGUGGGUGUG-UGGGUGGGCG-CAGCCGUUGGCGCUGGGCCGUAGAACGUGCCCAGGGCCCAGCGG---------CAAAUCGAACACACGCUUUUUGUCGCCACAGAGUGGCAA ....((((..((((((-((..(((...-..(((((((((.((((((((.....)).)))))).).)))))))---------)...)))..))))))))..)))).(((((...))))).. ( -58.40, z-score = -3.24, R) >droSec1.super_1 8518591 109 + 14215200 GCCAACAGUGGGUGUG-UGGGUGGGCG-CAGCCGUUGGCGCUGGGCCGUAGAACGUGCCCAGGGUCCAGCGG---------CAAAUCGAACACACGCUUUUUGUCGCCACAGAGUGUCAA ((..((((..((((((-((..(((...-..(((((((((.((((((((.....)).)))))).).)))))))---------)...)))..))))))))..)))).))............. ( -52.40, z-score = -2.42, R) >droYak2.chr2R 10944707 109 + 21139217 GCCACCAGUGGGUGUG-UGGGUGGGCG-CAGCCAUUGGCGCUGGGACGUAGAACGUGCCCAGGGCCCAGCGG---------CGAAACGAACACACGCUUUUUGUCGCCACGAAGUGGCAG (((((..(((((((((-((..((..((-(.((....(((.((((((((.....))).))))).)))..)).)---------))...))..))))))).........))))...))))).. ( -53.70, z-score = -2.27, R) >droEre2.scaffold_4845 7739288 102 - 22589142 -------GUGGGUGUG-UGGGUGGGCG-CAGCCAUUGGCGCUGGGCCGCAGAACGUGCCCAGGGCCCAGCGG---------CGAGACGAACACACGCUUUUUGUCGCCACGGCGUGGCAG -------..(((((((-((..((..((-(.((....(((.((((((((.....)).)))))).)))..)).)---------))...))..)))))))))......(((((...))))).. ( -50.40, z-score = -0.94, R) >droAna3.scaffold_13266 17560734 106 - 19884421 ------GCUAUUGGCGGUGGGUGGGCG-CAGCCGUUGGCGCUGGGCCAUAGAACGUGCCCAGGGCCCAGCAG----GCAACCACAAAGACCAAGA---UUUUGUCGCCACAUCUUCCCAA ------((.....))((.(((((((((-..(((((((((.(((((((.......).)))))).).))))).)----))....((((((.......---)))))))))).))))).))... ( -43.00, z-score = -0.67, R) >dp4.chr3 8891608 108 + 19779522 -------GUGGGGGCG-UGGGUGGGCG-CAG---UUGGCGCUGGGCCAUAGAACGUGCCCAGGGCCCACCACCACCACCACCACAACAACGAGGACCGUUUUGUCGCCACAAUGUGGCAA -------((((.((.(-..((((((((-(..---...)).(((((((.......).)))))).)))))))..).)).)))).((((.((((.....)))))))).(((((...))))).. ( -49.10, z-score = -0.69, R) >droPer1.super_4 2776985 108 + 7162766 -------GUGGGGGCG-UGGGUGGGCG-CAG---UUGGCGCUGGGCCAUAGAACGUGCCCAGGGCCCACCACCACCACCACCACAACAACGAGGACCGUUUUGUCGCCACAAUGUGGCAA -------((((.((.(-..((((((((-(..---...)).(((((((.......).)))))).)))))))..).)).)))).((((.((((.....)))))))).(((((...))))).. ( -49.10, z-score = -0.69, R) >droWil1.scaffold_180699 2440779 83 + 2593675 ------ACCGGGCGUG-GGUGUCUCCGUCAGUUGUUGGCUCGGGGCCAUAGAACGUUCCCAACAAAAAAAAA---AAAACCCGAAAGCAACAA--------------------------- ------.....((.((-(((...........(((((((..((...........))...))))))).......---...)))))...)).....--------------------------- ( -19.20, z-score = 0.89, R) >consensus ______AGUGGGUGUG_UGGGUGGGCG_CAGCCGUUGGCGCUGGGCCGUAGAACGUGCCCAGGGCCCAGCGG_________CAAAACGAACACACGCUUUUUGUCGCCACAGAGUGGCAA ...................(.(((((....((.....)).(((((((.......).)))))).))))).).................................................. (-18.35 = -18.80 + 0.45)


| Location | 11,024,303 – 11,024,394 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 66.00 |
| Shannon entropy | 0.63879 |
| G+C content | 0.43717 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.69 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
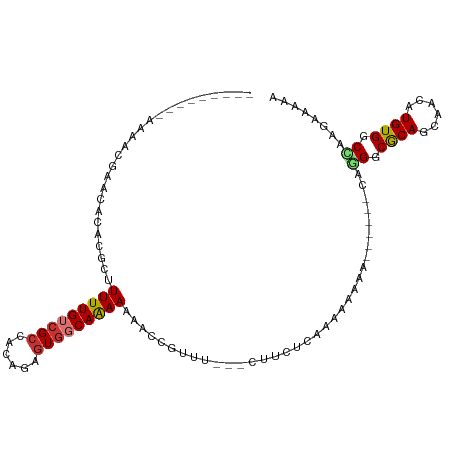
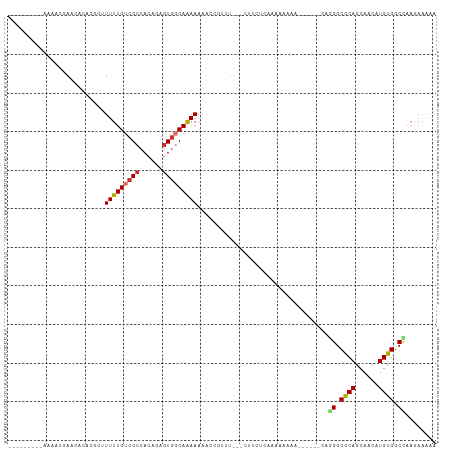
>dm3.chr2R 11024303 91 + 21146708 ---------AAAUCGAACACACGCUUUUUGUCGCCACAGAGUGGCAGAAUUAUCGUUU---CUUCUCGAAAAACA--------GGGCACAGCAAAAUGUGGCUAAGAAAAA ---------...((((....(((..(((((((((......)))))))))....)))..---....))))......--------((.((((......)))).))........ ( -19.70, z-score = -0.61, R) >droSim1.chr2R 9763078 93 + 19596830 ---------AAAUCGAACACACGCUUUUUGUCGCCACAGAGUGGCAAAAAAACCGUUU---CUUCUCAAAAAAAA------CAGGGCGCAGCAACAUGUGUCCAAGAAAAA ---------...(((((.(.(((.(((((...(((((...)))))..))))).))).)---.)))..........------..(((((((......)))))))..)).... ( -23.10, z-score = -2.10, R) >droSec1.super_1 8518662 99 + 14215200 ---------AAAUCGAACACACGCUUUUUGUCGCCACAGAGUGUCAAAAAAACUGUUC---AUUCUCAAAAAAAAAAAAAACAGGGCGCAGCAACAUGUGGCCAAGAAAAA ---------...(((((((.....((((((.(((......))).))))))...)))))---......................((.((((......)))).))..)).... ( -20.10, z-score = -1.25, R) >droYak2.chr2R 10944778 91 + 21139217 ---------GAAACGAACACACGCUUUUUGUCGCCACGAAGUGGCAGAAAAACCGUUU---CUUCUCGAAAAACA--------GGGCGCAGCAACAUGUGGCCAAGAAGAA ---------...........(((.(((((.(.(((((...)))))).))))).)))((---(((((.(.....).--------((.((((......)))).)).))))))) ( -23.30, z-score = -0.57, R) >droEre2.scaffold_4845 7739352 91 - 22589142 ---------GAGACGAACACACGCUUUUUGUCGCCACGGCGUGGCAGAAAAACCGUUU---CUUCUCAAAAAACA--------GGGCGCAGCAACAUGUGGCCAAGAAAAA ---------((((.(((...(((.(((((.(.(((((...)))))).))))).)))))---).))))........--------((.((((......)))).))........ ( -26.70, z-score = -1.37, R) >droAna3.scaffold_13266 17560800 73 - 19884421 ----CAACCACAAAGACCAAGA---UUUUGUCGCCACAUCUUCCCAAAAAAAUAA----------------------AAAACAAGGCGCAGCAACAUGUGGC--------- ----...(((((..(((.....---....)))(((....................----------------------.......))).........))))).--------- ( -10.13, z-score = -0.41, R) >dp4.chr3 8891669 111 + 19779522 ACCACCACCACAACAACGAGGACCGUUUUGUCGCCACAAUGUGGCAAAAAAAAAAUGUUGGCAUCUCUCUAACAAAAAAAUCAAGGCGCAGCAACAUGUGGCUAAAAGAGA .((((..((.(......).))...(((.(((((((((...)))))..........((((((.......))))))..........)))).))).....)))).......... ( -22.20, z-score = -0.05, R) >droPer1.super_4 2777046 111 + 7162766 ACCACCACCACAACAACGAGGACCGUUUUGUCGCCACAAUGUGGCAAAAAAAAAAUGUUGGCAUCUCUCUAACAAAAAAAUCAAGGCGCAGCAACAUGUGGCCAAAAGAGA .((((..((.(......).))...(((.(((((((((...)))))..........((((((.......))))))..........)))).))).....)))).......... ( -22.20, z-score = 0.12, R) >consensus _________AAAACGAACACACGCUUUUUGUCGCCACAGAGUGGCAAAAAAACCGUUU___CUUCUCAAAAAAAA______CAGGGCGCAGCAACAUGUGGCCAAGAAAAA .........................(((((((((......)))))))))..................................((.((((......)))).))........ (-13.48 = -13.69 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:17 2011