
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,898,214 – 2,898,345 |
| Length | 131 |
| Max. P | 0.568276 |

| Location | 2,898,214 – 2,898,308 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Shannon entropy | 0.30882 |
| G+C content | 0.44471 |
| Mean single sequence MFE | -22.14 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.516968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
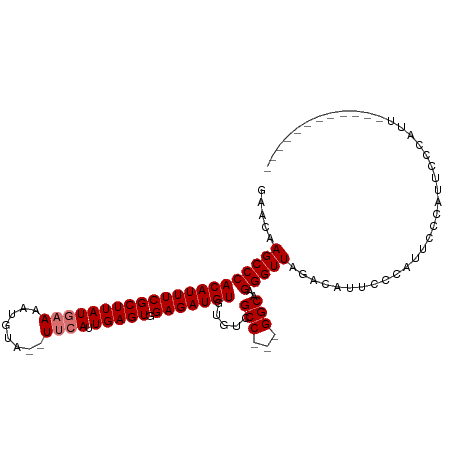
>dm3.chr2L 2898214 94 + 23011544 GAACAAGCCCACAUUUCGCUUAUGAAAAUGUA--UUCAUUGAGUGGAGAUGUAUGUCGCCGCCGGCAAGGGUU--CCAUUCCCAUUGCCAUUUCCAUU------------ ..........((((((((((((((((......--)))).))))).)))))))...........((((((((..--.....))).))))).........------------ ( -24.50, z-score = -1.27, R) >droSec1.super_5 1056894 110 + 5866729 AAACAAGCCCACAUUUCGCUUAUGAAAAUAAAAGUUCAUUGAGUGGAGAUGUUUUUCGCCACCGGCAAGGGUUAGACAUUCCCAUUCCCAUUCCCGUUGCCAUUUCCAUU ......((..((((((((((((((((........)))).))))).))))))).....))....((((((((.....................))).)))))......... ( -22.40, z-score = -0.92, R) >droYak2.chr2L 2893439 87 + 22324452 GAACAAGCCCACAUUUCGCUUAUGAAAAUGUA--UGCAUUGAGUGGAGAUGUGUGUCGCC---GGCAAGGGUUAGACAUUCCCAUUCCCAUU------------------ ......((..((((((((....)).)))))).--.))...((((((..((((.((.(.((---.....))).)).))))..)))))).....------------------ ( -21.60, z-score = -0.54, R) >droEre2.scaffold_4929 2945590 99 + 26641161 GAACAAGCCCACAUUUCGCUUAUAAAAAUGUA--UUCAUUGAGUGGAGAUGUGUGUCGCC---GGCAAGGGUUAGACAUUCCCAUUCUCAUACCCAUUCCCAUU------ ......((((((((((((((((..........--.....))))).)))))))).......---)))..((((.(((.........)))...)))).........------ ( -20.07, z-score = -0.27, R) >consensus GAACAAGCCCACAUUUCGCUUAUGAAAAUGUA__UUCAUUGAGUGGAGAUGUGUGUCGCC___GGCAAGGGUUAGACAUUCCCAUUCCCAUUCCCAUU____________ .....(((((((((((((((((((((........)))).))))).))))))).....(((...)))..)))))..................................... (-19.60 = -20.10 + 0.50)


| Location | 2,898,249 – 2,898,345 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.00 |
| Shannon entropy | 0.29064 |
| G+C content | 0.42690 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -18.87 |
| Energy contribution | -18.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2898249 96 + 23011544 AUUGAGUGGAGAUGUAUGUCGCCGCCGGCAAGGGUU--------------CCAUUCCCAUUGCCAUUUCCAUUUCCAUUCUCGUAAAUACAAAACAAGAAAAAUGAGUCG ...(((((((((((...((....)).((((((((..--------------.....))).))))).....))))))))))).............................. ( -24.80, z-score = -2.09, R) >droSec1.super_5 1056931 110 + 5866729 AUUGAGUGGAGAUGUUUUUCGCCACCGGCAAGGGUUAGACAUUCCCAUUCCCAUUCCCGUUGCCAUUUCCAUUUCCAUUCUCGUAAAUACAAAACAAGAAAAAUGAGUGG ...(((((((((((......(((...)))..(((.........))).................)))))))))))((((((........................)))))) ( -23.36, z-score = -0.57, R) >droYak2.chr2L 2893474 89 + 22324452 AUUGAGUGGAGAUGUGUGUCGCC---GGCAAGGGUUAGACAU------UCCCAUUC------------CCAUUUCCAUUCUCGUAAAUACAAAACAAGAAAAAUGAGUGG ...((((((..((((.((.(.((---.....))).)).))))------..))))))------------((((((...((((.((.........)).))))....)))))) ( -22.20, z-score = -1.28, R) >droEre2.scaffold_4929 2945625 101 + 26641161 AUUGAGUGGAGAUGUGUGUCGCC---GGCAAGGGUUAGACAU------UCCCAUUCUCAUACCCAUUCCCAUUUUCAUUCUCGUAAAUACAAAACAAGAAAAAUGAGUGG ...((((((..((((.((.(.((---.....))).)).))))------..))))))......((((((...(((((.....................)))))..)))))) ( -23.50, z-score = -0.96, R) >consensus AUUGAGUGGAGAUGUGUGUCGCC___GGCAAGGGUUAGACAU______UCCCAUUCCCAUUGCCAUUUCCAUUUCCAUUCUCGUAAAUACAAAACAAGAAAAAUGAGUGG ...(((((((((((......(((...)))..(((...............))).................))))))))))).............................. (-18.87 = -18.93 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:34 2011