
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,985,522 – 10,985,612 |
| Length | 90 |
| Max. P | 0.773041 |

| Location | 10,985,522 – 10,985,612 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 76.33 |
| Shannon entropy | 0.40065 |
| G+C content | 0.50425 |
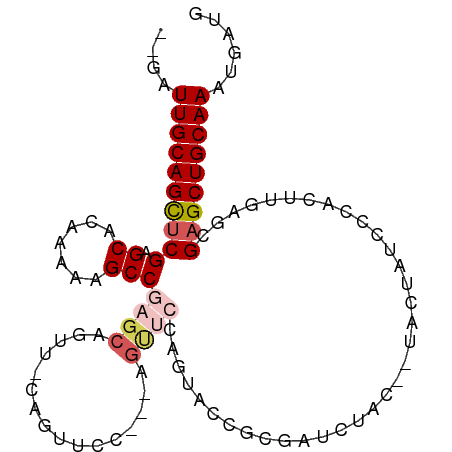
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.64 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
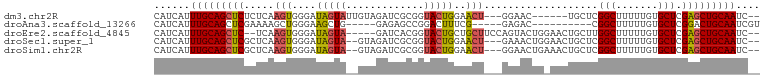
>dm3.chr2R 10985522 90 + 21146708 CAUCAUUUGCAGCUCUCUCAAGUGGGAUAGUAUUGUAGAUCGCGGUACUGGAACU---GGAAC------UGCUCGGCUUUUUGUGCUCGAGCUGCAAUC-- ......(((((((((((((....)))).(((...((((.((.((((......)))---))).)------)))...)))..........)))))))))..-- ( -28.80, z-score = -1.67, R) >droAna3.scaffold_13266 17530333 81 - 19884421 CAUCAUUUGCAGCUCGAAAAGCUGGGAAGCUG-----GAGAGCCGGACUUUCG-----GAGAC----------CGGCUUUUUGUGCUCGGACUGCAAUCGU ......((((((..(((..((((....))))(-----(((((((((.(((...-----))).)----------)))))))))....)))..)))))).... ( -31.90, z-score = -2.20, R) >droEre2.scaffold_4845 7708401 92 - 22589142 CAUCAUUUGCAGCUC--UCAAGUGGGAUAGUA-----GAUCACGGUACUGCUGCUUCCAGUACUGGAACUGCUUGGCUUUUUGUGCUCGAGCUGCAAUC-- ......(((((((((--.((((..((((....-----.))).((((((((.......))))))))...)..))))((.......))..)))))))))..-- ( -34.70, z-score = -2.92, R) >droSec1.super_1 8488279 94 + 14215200 CAUCAUUUGCAGCUCGCUCAAGUGGGAUAGUA--GUAGAUCGCGGUACUGGAACU---GAAACUGGAACUGCUCGGCUUUUUGUGCUCGAGCUGCAAUC-- ......((((((((((....((((.((....(--((.((..(((((.(..(....---....)..).))))))).)))..)).))))))))))))))..-- ( -30.40, z-score = -1.43, R) >droSim1.chr2R 9730774 94 + 19596830 CAUCAUUUGCAGCUCGCUCAAGUGGGAUAGUA--GUAGAUCGCGGUACUGGAACU---GGAACUGAAACUGCUCGGCUUUUUGUGCUCGAGCUGCAAUC-- ......(((((((((((.(((((.(..((((.--.(((.((.((((......)))---))).)))..))))..).)))...)).))..)))))))))..-- ( -32.60, z-score = -2.15, R) >consensus CAUCAUUUGCAGCUCGCUCAAGUGGGAUAGUA__GUAGAUCGCGGUACUGGAACU___GGAACUG_AACUGCUCGGCUUUUUGUGCUCGAGCUGCAAUC__ ......(((((((((......((((..............))))...............................(((.......))).))))))))).... (-15.16 = -15.64 + 0.48)


| Location | 10,985,522 – 10,985,612 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.33 |
| Shannon entropy | 0.40065 |
| G+C content | 0.50425 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -14.02 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
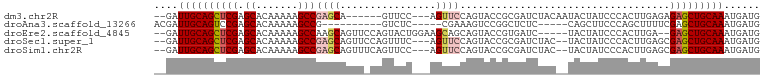
>dm3.chr2R 10985522 90 - 21146708 --GAUUGCAGCUCGAGCACAAAAAGCCGAGCA------GUUCC---AGUUCCAGUACCGCGAUCUACAAUACUAUCCCACUUGAGAGAGCUGCAAAUGAUG --..((((((((((.((.......))))))).------.....---............((..(((.(((...........))))))..)))))))...... ( -20.50, z-score = -1.25, R) >droAna3.scaffold_13266 17530333 81 + 19884421 ACGAUUGCAGUCCGAGCACAAAAAGCCG----------GUCUC-----CGAAAGUCCGGCUCUC-----CAGCUUCCCAGCUUUUCGAGCUGCAAAUGAUG .((.((((((..((((.......(((((----------(.((.-----....)).))))))...-----.((((....)))).))))..)))))).))... ( -28.70, z-score = -3.22, R) >droEre2.scaffold_4845 7708401 92 + 22589142 --GAUUGCAGCUCGAGCACAAAAAGCCAAGCAGUUCCAGUACUGGAAGCAGCAGUACCGUGAUC-----UACUAUCCCACUUGA--GAGCUGCAAAUGAUG --..((((((((((.((.......))).((.((.(((.((((((.......)))))).).)).)-----).))...........--)))))))))...... ( -26.40, z-score = -1.77, R) >droSec1.super_1 8488279 94 - 14215200 --GAUUGCAGCUCGAGCACAAAAAGCCGAGCAGUUCCAGUUUC---AGUUCCAGUACCGCGAUCUAC--UACUAUCCCACUUGAGCGAGCUGCAAAUGAUG --..(((((((((..((.(........).)).((((.(((...---(((...((((........)))--)))).....))).)))))))))))))...... ( -23.50, z-score = -1.33, R) >droSim1.chr2R 9730774 94 - 19596830 --GAUUGCAGCUCGAGCACAAAAAGCCGAGCAGUUUCAGUUCC---AGUUCCAGUACCGCGAUCUAC--UACUAUCCCACUUGAGCGAGCUGCAAAUGAUG --..(((((((((((((..(((..((...))..)))..)))).---.((((.(((...(.(((....--....)))).))).)))))))))))))...... ( -23.80, z-score = -1.43, R) >consensus __GAUUGCAGCUCGAGCACAAAAAGCCGAGCAGUU_CAGUUCC___AGUUCCAGUACCGCGAUCUAC__UACUAUCCCACUUGAGCGAGCUGCAAAUGAUG ....((((((((((.((.......)))((((.......))))............................................)))))))))...... (-14.02 = -14.90 + 0.88)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:12 2011