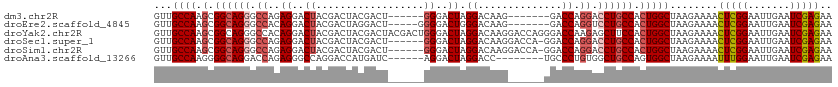
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,970,532 – 10,970,632 |
| Length | 100 |
| Max. P | 0.992836 |

| Location | 10,970,532 – 10,970,632 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 85.49 |
| Shannon entropy | 0.26196 |
| G+C content | 0.55214 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -22.40 |
| Energy contribution | -22.79 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
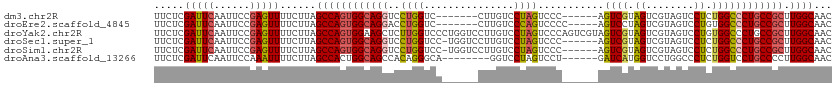
>dm3.chr2R 10970532 100 + 21146708 UUCUCGAUUCAAUUCCGAGUUUUCUUAGCCAGUGGCAGGUCCUGGUC-------CUUGUCCUAGUCCC------AGUCGUAGUCGUAGUCCUCUGGCCCUGCCGCUUGGCAAC ..((((.........))))........((((((((((((..((((..-------((......))..))------)).....((((........)))))))))))).))))... ( -34.10, z-score = -3.90, R) >droEre2.scaffold_4845 7699142 101 - 22589142 UUCUCGAUUCAAUUCCGAGUUUUCUUAGCCAGUGGCAGGACCUGGUC-------CUUGUCCCAGUCCCC-----AGUCCUAGUCGUAGUCCUGUGGCCCUGCCGCUUGGCAAC ..((((.........))))........((((((((((((..((((..-------......))))..(((-----((..(((....)))..))).)).)))))))).))))... ( -37.10, z-score = -4.25, R) >droYak2.chr2R 10905044 113 + 21139217 UUCUCGAUUCAAUUCCGAGUUUUCUUAGCCAGUGGAAGCUCUUGGUCCCUGGUCCUUGUCCUAGUCCCAGUCGUAGUCGUAGUCGUAGUCCUGUGGCCCUGCCGCUUGGCAAC ..((((((((......((((((((.........)))))))).......((((..((......))..))))..).))))).)).....(((..(((((...)))))..)))... ( -29.80, z-score = -1.26, R) >droSec1.super_1 8480267 106 + 14215200 UUCUCGAUUCAAUUCCGAGUUUUCUUAGCCAGUGGCAGGUCCUGGUCC-UGGUCCUUGUCCUAGUCCC------AGUCGUAGUCGUAGUCCUCUGGCCCUGCCGCUUGGCAAC ..((((.........))))........((((((((((((.((.((.((-(((..((......))..))------)).((....))..).))...)).)))))))).))))... ( -37.70, z-score = -4.19, R) >droSim1.chr2R 9722776 106 + 19596830 UUCUCGAUUCAAUUCCGAGUUUUCUUAGCCAGUGGCAGGUCCUGGUCC-UGGUCCUUGUCCUAGUCCC------AGUCGUAGUCGUAGUCCUCUGGCCCUGCCGCUUGGCAAC ..((((.........))))........((((((((((((.((.((.((-(((..((......))..))------)).((....))..).))...)).)))))))).))))... ( -37.70, z-score = -4.19, R) >droAna3.scaffold_13266 17521995 99 - 19884421 UUCUCGAUUCAAUUCCAAAUUUUCUUAGCCACUGGCAGCCACAGGGCA--------GGUCCUAGUCCU------GAUCAUGGUCCUGGCCCUCUGGUCCUGCCCCUUGGCAAC ...........................((((..((((((((.((((((--------((.((.......------......)).))).))))).)))..)))))...))))... ( -34.52, z-score = -2.40, R) >consensus UUCUCGAUUCAAUUCCGAGUUUUCUUAGCCAGUGGCAGGUCCUGGUCC______CUUGUCCUAGUCCC______AGUCGUAGUCGUAGUCCUCUGGCCCUGCCGCUUGGCAAC .....(((((......)))))......((((((((((((..((((...............))))...........((((.((........)).)))))))))))).))))... (-22.40 = -22.79 + 0.39)
| Location | 10,970,532 – 10,970,632 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.49 |
| Shannon entropy | 0.26196 |
| G+C content | 0.55214 |
| Mean single sequence MFE | -35.64 |
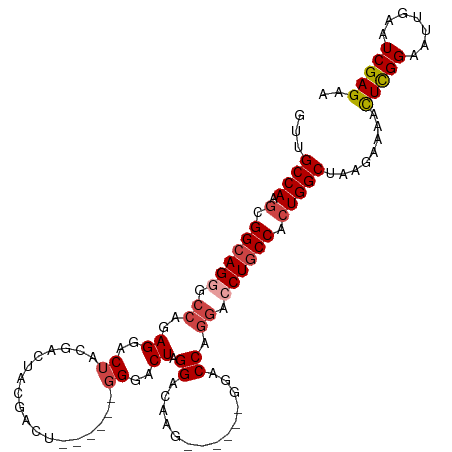
| Consensus MFE | -20.04 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
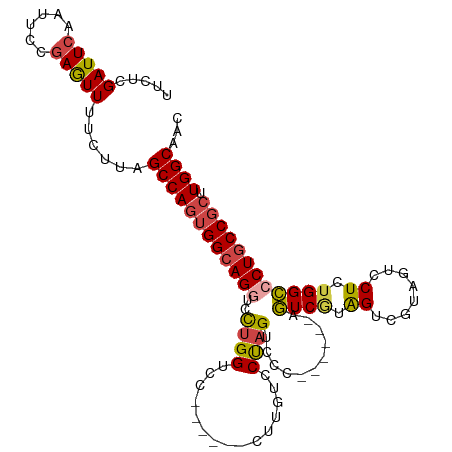
>dm3.chr2R 10970532 100 - 21146708 GUUGCCAAGCGGCAGGGCCAGAGGACUACGACUACGACU------GGGACUAGGACAAG-------GACCAGGACCUGCCACUGGCUAAGAAAACUCGGAAUUGAAUCGAGAA ...((((.(.((((((.....((........))....((------((..((......))-------..))))..)))))).)))))........(((((.......))))).. ( -32.60, z-score = -2.53, R) >droEre2.scaffold_4845 7699142 101 + 22589142 GUUGCCAAGCGGCAGGGCCACAGGACUACGACUAGGACU-----GGGGACUGGGACAAG-------GACCAGGUCCUGCCACUGGCUAAGAAAACUCGGAAUUGAAUCGAGAA ...((((.(.(((((((((.(((..(((....)))..))-----)((..((......))-------..)).))))))))).)))))........(((((.......))))).. ( -43.70, z-score = -4.65, R) >droYak2.chr2R 10905044 113 - 21139217 GUUGCCAAGCGGCAGGGCCACAGGACUACGACUACGACUACGACUGGGACUAGGACAAGGACCAGGGACCAAGAGCUUCCACUGGCUAAGAAAACUCGGAAUUGAAUCGAGAA .(((((....)))))(((((..(((...((....)).......((((..((......))..))))............)))..))))).......(((((.......))))).. ( -28.80, z-score = -0.51, R) >droSec1.super_1 8480267 106 - 14215200 GUUGCCAAGCGGCAGGGCCAGAGGACUACGACUACGACU------GGGACUAGGACAAGGACCA-GGACCAGGACCUGCCACUGGCUAAGAAAACUCGGAAUUGAAUCGAGAA ...((((.(.((((((.((...((....((....)).((------((..((......))..)))-)..)).)).)))))).)))))........(((((.......))))).. ( -35.90, z-score = -2.85, R) >droSim1.chr2R 9722776 106 - 19596830 GUUGCCAAGCGGCAGGGCCAGAGGACUACGACUACGACU------GGGACUAGGACAAGGACCA-GGACCAGGACCUGCCACUGGCUAAGAAAACUCGGAAUUGAAUCGAGAA ...((((.(.((((((.((...((....((....)).((------((..((......))..)))-)..)).)).)))))).)))))........(((((.......))))).. ( -35.90, z-score = -2.85, R) >droAna3.scaffold_13266 17521995 99 + 19884421 GUUGCCAAGGGGCAGGACCAGAGGGCCAGGACCAUGAUC------AGGACUAGGACC--------UGCCCUGUGGCUGCCAGUGGCUAAGAAAAUUUGGAAUUGAAUCGAGAA ...((((...(((((..(((.(((((.(((.((......------.......)).))--------)))))).))))))))..))))........(((.((......)).))). ( -36.92, z-score = -1.87, R) >consensus GUUGCCAAGCGGCAGGGCCAGAGGACUACGACUACGACU______GGGACUAGGACAAG______GGACCAGGACCUGCCACUGGCUAAGAAAACUCGGAAUUGAAUCGAGAA ...((((.(.((((((.((..((..((..................))..)).((..............)).)).)))))).)))))........(((((.......))))).. (-20.04 = -20.93 + 0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:11 2011