
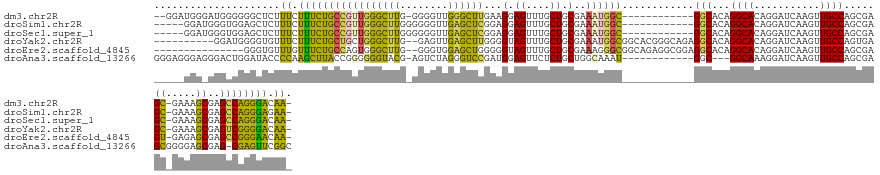
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,966,636 – 10,966,762 |
| Length | 126 |
| Max. P | 0.662642 |

| Location | 10,966,636 – 10,966,762 |
|---|---|
| Length | 126 |
| Sequences | 6 |
| Columns | 143 |
| Reading direction | reverse |
| Mean pairwise identity | 69.05 |
| Shannon entropy | 0.55958 |
| G+C content | 0.60302 |
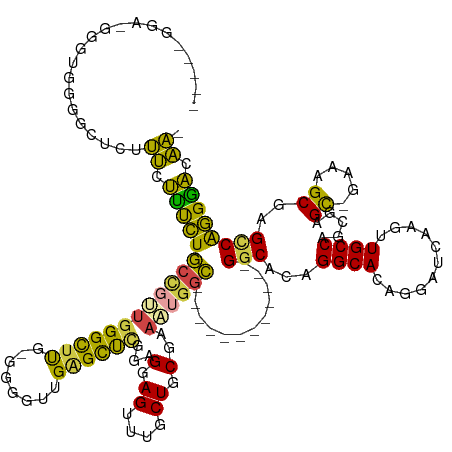
| Mean single sequence MFE | -48.22 |
| Consensus MFE | -22.49 |
| Energy contribution | -21.84 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10966636 126 - 21146708 --GGAUGGGAUGGGGGGCUCUUUCUUUCUGCCGUUGGGCUUG-GGGGUUGGGCUUGAAGGAGUUUGCUGCGAAAUGGC------------GGCACAGGCACAGGAUCAAGUUGCCAGCGAGC-GAAAGCGAGCCAGGGACAA- --.............(((((...(((((.(((((((((((((-(...(((.(((((........((((((......))------------))))))))).)))..))))))..)))))).))-))))).)))))........- ( -46.00, z-score = -0.58, R) >droSim1.chr2R 9718882 124 - 19596830 -----GGAUGGGUGGAGCUCUUUCUUUCUGCCGUUGGGCUUGGGGGGUUGAGCUCGGAGGAGUUUGCUGCGAAAUGGC------------GGCACAGGCACAGGAUCAAGUUGCCAGCGAGC-GAAAGCGAGCCAGGGAGAA- -----.(((..(((((((((..(((..(.(((....)))..)..)))..)))))).........((((((......))------------))))....)))...)))...((.((.((..((-....))..))..)).))..- ( -46.30, z-score = -0.47, R) >droSec1.super_1 8476386 124 - 14215200 -----GGAUGGGUGGAGCUCUUUCUUUCUGCCGUUGGGCUUGGGGGGUUGAGCUCGGAGGAGUUUGCUGCGAAAUGGC------------GGCACAGGCACAGGAUCAAGUUGCCAGCGAGC-GAAAGCGAGCCAGGGACAA- -----.(((..(((((((((..(((..(.(((....)))..)..)))..)))))).........((((((......))------------))))....)))...)))..(((.(..((..((-....))..))..).)))..- ( -48.30, z-score = -1.02, R) >droYak2.chr2R 10901053 129 - 21139217 ----------GGAUGGGGUGUUUCUUUCUGCUGCUGGGCUUG--GAGUUGAGCUUGGGGUAGUUUGCUGCGAAAUGGCGGCACGGGCAGAGGCACAGGCACAGGAUCAAGUUGCCAGUGAGC-GAAAGCGAGUCGGGGACAA- ----------.(((...((((((.((((((((.(..(((((.--.....)))))..).......((((((......))))))..))))))))...))))))...)))..(((.((..(..((-....))..)..)).)))..- ( -47.30, z-score = -1.27, R) >droEre2.scaffold_4845 7695365 124 + 22589142 ---------------GGGUGUUUGUUUCUGCCAGUGGGCUUG--GGGUGGAGCUGGGGGUAGUUUGCUGCGAAAGGGCGGCAGAGGCGGAGGCACAGGCACAGGAUCAAGUUGCCAGCGAGU-GAGAGCGAGCCGGGAACAA- ---------------..(((((((((((((((..(.(((((.--.....))))).)......((((((((......))))))))))))))))))..)))))........(((.((.((..((-....))..)).)).)))..- ( -49.70, z-score = -2.47, R) >droAna3.scaffold_13266 17517422 126 + 19884421 GGGAGGGAGGGACUGGAUACCCCAAGCUUACCGGGGGGUACG-AGUCUAGGGUCCGAUGGAGUUCUCUGCUGGCAAAU------------GGC---GGCAAAGGAUCAAGUUGCCAGCGAGCGGGGAGCGAG-CGAGUUCGGC ....(((..(((((.(.((((((..(.....)..))))))).-)))))....)))......((((((((((.((...(------------(((---(((..........))))))))).))))))))))(((-....)))... ( -51.70, z-score = -1.86, R) >consensus _____GGA_GGGUGGGGCUCUUUCUUUCUGCCGUUGGGCUUG_GGGGUUGAGCUCGGAGGAGUUUGCUGCGAAAUGGC____________GGCACAGGCACAGGAUCAAGUUGCCAGCGAGC_GAAAGCGAGCCAGGGACAA_ ........................((((((((.((((((((........)))))))).)).(((((((((....((((.............((....)).............))))....))....))))))))))))).... (-22.49 = -21.84 + -0.66)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:09 2011