| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,954,716 – 10,954,819 |
| Length | 103 |
| Max. P | 0.962075 |

| Location | 10,954,716 – 10,954,819 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 69.76 |
| Shannon entropy | 0.64858 |
| G+C content | 0.43553 |
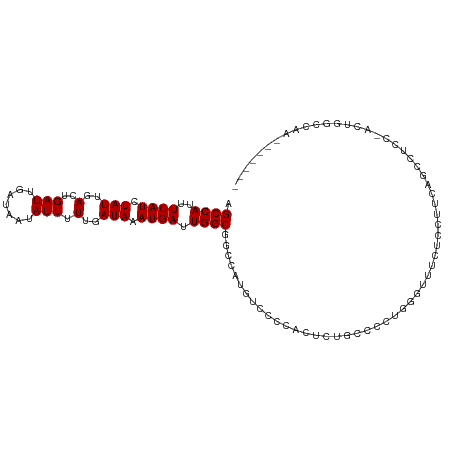
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.65 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.962075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
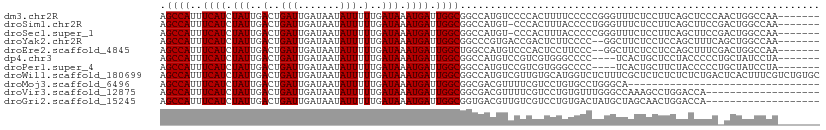
>dm3.chr2R 10954716 103 - 21146708 AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCGGCCAUGUCCCCACUUUUCCCCCGGGUUUCUCCUUCAGCUCCCAACUGGCCAA------- .((((..((((.(((..(..(((.......))).)..))).)))).))))(((((.((....)).........(((...((.....))..)))...)))))..------- ( -22.90, z-score = -1.02, R) >droSim1.chr2R 9707061 102 - 19596830 AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCGGCCAUGU-CCCACUUUACCCCUGGGUUUCUCCUUCAGCUUCCGACUGGCCAA------- .((((..((((.(((..(..(((.......))).)..))).)))).))))(((((.((-(.......(((....)))................))))))))..------- ( -25.10, z-score = -1.81, R) >droSec1.super_1 8464492 102 - 14215200 AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCGGCCAUGU-CCCACUUUACCCCCGGGUUUCUCCUUCAGCUUCCGACUGGCCAA------- .((((..((((.(((..(..(((.......))).)..))).)))).))))(((((.((-(.......(((....)))................))))))))..------- ( -25.10, z-score = -1.79, R) >droYak2.chr2R 10888903 101 - 21139217 AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCGGCCCGUGACCGACUCUUCCCC--GGCUUCUCCUCCAGCUUUCAGCUGGCCAA------- .((((..((((.(((..(..(((.......))).)..))).)))).))))((((.(.((........)).).--))))......(((((.....)))))....------- ( -24.00, z-score = -1.27, R) >droEre2.scaffold_4845 7683645 101 + 22589142 AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCUGGCCAUGUCCCACUCCUUCCC--GGCUUCUCCUCCAGCUUUCGACUGGCCAA------- (((((..((((.(((..(..(((.......))).)..))).)))).)))))(((((.(((............--((((........))))...))))))))..------- ( -24.26, z-score = -1.99, R) >dp4.chr3 8841215 99 - 19779522 AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCGGCCAUGUCCGUCGUGGGCCCC----UCACUGCUCCUACCCCCUGCUAUCCUA------- .((((..((((.(((..(..(((.......))).)..))).)))).))))(((((((.....))).))))..----...........................------- ( -19.80, z-score = -0.52, R) >droPer1.super_4 2726216 99 - 7162766 AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCGGCCAUGUCCGUCGUGGGCCCC----UCACUGCUUCUACCCCCUGCUAUCCUA------- .((((..((((.(((..(..(((.......))).)..))).)))).))))(((((((.....))).))))..----...........................------- ( -19.80, z-score = -0.52, R) >droWil1.scaffold_180699 2373720 110 - 2593675 AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCGGCCAUGUCGUUGUGCAUGGUCUCUUUCGCUCUCUCUCUCUGACUCACUUUCGUCUGUGC .((((..((((.(((..(..(((.......))).)..))).)))).))))((((((((......))))))))...................(((........)))..... ( -24.50, z-score = -1.46, R) >droMoj3.scaffold_6496 9642395 78 + 26866924 AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCGGCGACGUUUUCGUCCUGUGCCUGGGCA-------------------------------- .((((..((((.(((..(..(((.......))).)..))).)))).))))(((((((....))))....)))......-------------------------------- ( -19.20, z-score = -1.12, R) >droVir3.scaffold_12875 9823510 91 - 20611582 AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCGGCGACGUUUUCGUCCUGUGUUUGGGCCAAAGCCUGGACCA------------------- .((((..((((.(((..(..(((.......))).)..))).)))).))))((.((((....))))))(.(((..(((....)))..)))).------------------- ( -27.00, z-score = -2.38, R) >droGri2.scaffold_15245 3028831 91 - 18325388 AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCGGUGACGUUGUCGUCCUGUGACUAUGCUAGCAACUGGACCA------------------- .((((..((((.(((..(..(((.......))).)..))).)))).))))(....)......((((.((..(((...)))..)).))))..------------------- ( -20.70, z-score = -1.10, R) >consensus AGCCAUUUCAUCUAUUGACUGAUUGAUAAUAUUUUUGAUAAAUGAUUGGCGGCCAUGUCCCCACUCUGCCCCUGGGUUUCUCCUUCAGCCUCC_ACUGGCCAA_______ .((((..((((.(((..(..(((.......))).)..))).)))).))))............................................................ (-11.65 = -11.65 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:08 2011