| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,950,083 – 10,950,308 |
| Length | 225 |
| Max. P | 0.801066 |

| Location | 10,950,083 – 10,950,211 |
|---|---|
| Length | 128 |
| Sequences | 12 |
| Columns | 159 |
| Reading direction | forward |
| Mean pairwise identity | 72.96 |
| Shannon entropy | 0.50150 |
| G+C content | 0.34803 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -14.17 |
| Energy contribution | -14.04 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
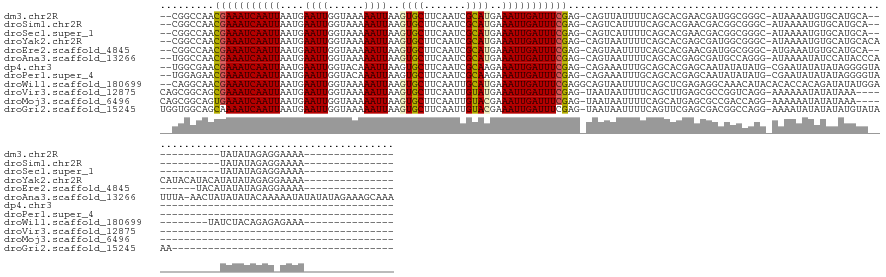
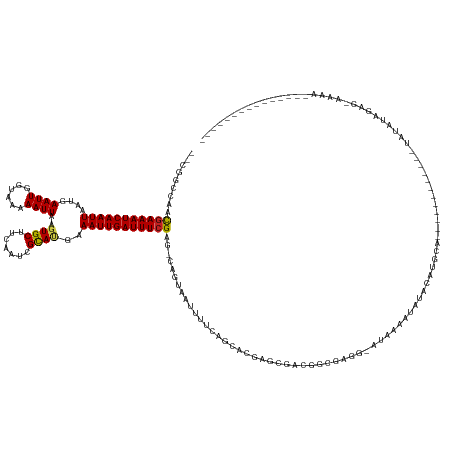
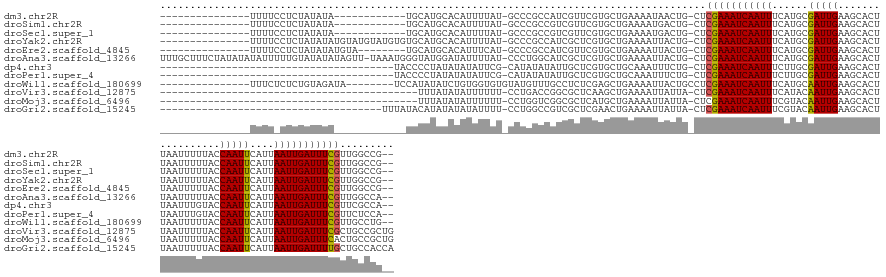
>dm3.chr2R 10950083 128 + 21146708 --CGGCCAACGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUCGCAUGAAAUUGAUUUCGAG-CAGUUAUUUUCAGCACGAACGAUGGCGGGC-AUAAAAUGUGCAUGCA------------UAUAUAGAGGAAAA--------------- --...((..(((((((((((....((((......))))..((((.......))))..))))))))))).(-(((((((((((.....))).))))))..((-((.....)))).))).------------........))....--------------- ( -28.10, z-score = -0.64, R) >droSim1.chr2R 9702402 128 + 19596830 --CGGCCAACGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUCGCAUGAAAUUGAUUUCGAG-CAGUCAUUUUCAGCACGAACGACGGCGGGC-AUAAAAUGUGCAUGCA------------UAUAUAGAGGAAAA--------------- --..(((..(((((((((((....((((......))))..((((.......))))..))))))))))).(-(.(((...(((.....))).))).)).)))-.....(((((....))------------)))...........--------------- ( -28.70, z-score = -0.70, R) >droSec1.super_1 8459833 128 + 14215200 --CGGCCAACGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUCGCAUGAAAUUGAUUUCGAG-CAGUCAUUUUCAGCACGAACGACGGCGGGC-AUAAAAUGUGCAUGCA------------UAUAUAGAGGAAAA--------------- --..(((..(((((((((((....((((......))))..((((.......))))..))))))))))).(-(.(((...(((.....))).))).)).)))-.....(((((....))------------)))...........--------------- ( -28.70, z-score = -0.70, R) >droYak2.chr2R 10884246 140 + 21139217 --CGGCCAACGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUCGCAUGAAAUUGAUUUCGAG-CAGUAAUUUUCAGCACGAGCGAUGGCGGGC-AUAAAAUGUGCAUGCACACAUACAUACAUAUAUAGAGGAAAA--------------- --(.((((.(((((((((((....((((......))))..((((.......))))..)))))))))))..-............((....))..)))).)..-.....(((((......))))).....................--------------- ( -27.40, z-score = 0.14, R) >droEre2.scaffold_4845 7679274 132 - 22589142 --CGGCCAACGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUCGCAUGAAAUUGAUUUCGAG-CAGUAAUUUUCAGCACGAACGAUGGCGGGC-AUGAAAUGUGCAUGCA--------UACAUAUAUAGAGGAAAA--------------- --...((..(((((((((((....((((......))))..((((.......))))..))))))))))).(-(((((..(((((((.((........)).))-.)))))..))).))).--------............))....--------------- ( -29.60, z-score = -0.79, R) >droAna3.scaffold_13266 17500731 154 - 19884421 --UGGCCAACGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUCGCAUGAAAUUGAUUUCGAG-CAGUAAUUUUCAGCACGAGCGAUGCCAGGG-AUAAAAUAUCCAUACCCAUUUA-AACUAUAUAUACAAAAAUAUAUAUAGAAAGCAAA --((((...(((((((((((....((((......))))..((((.......))))..)))))))))))..-............((....))...)))).((-(((...)))))...........-..(((((((((......)))))))))........ ( -32.00, z-score = -2.44, R) >dp4.chr3 8837313 116 + 19779522 --UGGCGAACGAAAUCAAUUAAUGAAUUGGUACAAAUUAAGUGCUUCAAUCGCAAGAAAUUGAUUUCGAG-CAGAAAUUUGCAGCACGAGCAAUAUAUAUG-CGAAUAUAUAUAGGGGUA--------------------------------------- --..((...(((((((((((..(((...(((((.......)))))))).((....))))))))))))).(-(((....)))).))....((..((((((((-....))))))))...)).--------------------------------------- ( -29.50, z-score = -3.02, R) >droPer1.super_4 2722319 116 + 7162766 --UGGAGAACGAAAUCAAUUAAUGAAUUGGUACAAAUUAAGUGCUUCAAUCGCAAGAAAUUGAUUUCGAG-CAGAAAUUUGCAGCACGAGCAAUAUAUAUG-CGAAUAUAUAUAGGGGUA--------------------------------------- --.......(((((((((((..(((...(((((.......)))))))).((....))))))))))))).(-(((....)))).((....)).((((((((.-...)))))))).......--------------------------------------- ( -28.00, z-score = -3.06, R) >droWil1.scaffold_180699 2369212 134 + 2593675 --CAGGCAACGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUUGCAUGAAAUUGAUUUCGAGGCAGUAAUUUUCAGCUCGAGAGGCAAACAUACACACCACAGAUAUAUGGA--------UAUCUACAGAGAGAAA--------------- --...((..(((((((((((.(((((((((...............)))))).)))..)))))))))))..)).....(((((.((.(....)))...............((((((....)--------))))).....))))).--------------- ( -30.16, z-score = -2.56, R) >droVir3.scaffold_12875 9818769 114 + 20611582 CAGCGGCAGCGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUUGUAUGAAAUUGAUUUCGAG-UAAUAAUUUUCAGCUUGAGCGCCGGUCAGG-AAAAAAUAUAUAAA------------------------------------------- ...((((..(((((((((((.(((((((((...............)))))).)))..)))))))))))..-............((....))))))......-..............------------------------------------------- ( -23.56, z-score = -1.11, R) >droMoj3.scaffold_6496 9636982 114 - 26866924 CAGCGGCAGUGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUUGUACGAAAUUGAUUUCGAG-UAAUAAUUUUCAGCAUGAGCGCCGACCAGG-AAAAAAUAUAUAAA------------------------------------------- ...((((..(((((((((((....((((......))))..((((.......))))..)))))))))))..-............((....))))))......-..............------------------------------------------- ( -22.70, z-score = -1.33, R) >droGri2.scaffold_15245 3024299 120 + 18325388 UGGUGGCAGCAAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUUGUACGAAAUUGAUUUCGAG-UAAUAAUUUUCAGUUCGAGCGACGGCCAGG-AAAAUAUAUAUAUGUAUAAA------------------------------------- ((((.(..((..((((((((....((((......))))..((((.......))))..))))))))(((((-.............)))))))..).))))..-....(((((....)))))..------------------------------------- ( -21.22, z-score = -0.35, R) >consensus __CGGCCAACGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUCGCAUGAAAUUGAUUUCGAG_CAGUAAUUUUCAGCACGAGCGACGGCGAGG_AUAAAAUAUACAUGCA____________UAUAUAGAG_AAAA_______________ .........(((((((((((....((((......))))..((((.......))))..)))))))))))........................................................................................... (-14.17 = -14.04 + -0.12)
| Location | 10,950,083 – 10,950,211 |
|---|---|
| Length | 128 |
| Sequences | 12 |
| Columns | 159 |
| Reading direction | reverse |
| Mean pairwise identity | 72.96 |
| Shannon entropy | 0.50150 |
| G+C content | 0.34803 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -11.00 |
| Energy contribution | -10.79 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10950083 128 - 21146708 ---------------UUUUCCUCUAUAUA------------UGCAUGCACAUUUUAU-GCCCGCCAUCGUUCGUGCUGAAAAUAACUG-CUCGAAAUCAAUUUCAUGCGAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGUUGGCCG-- ---------------..............------------.(((((.......)))-))..((((.(....(.((...........)-).)((((((((((..(((.(((((.................)))))))).))))))))))).))))..-- ( -22.33, z-score = -0.63, R) >droSim1.chr2R 9702402 128 - 19596830 ---------------UUUUCCUCUAUAUA------------UGCAUGCACAUUUUAU-GCCCGCCGUCGUUCGUGCUGAAAAUGACUG-CUCGAAAUCAAUUUCAUGCGAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGUUGGCCG-- ---------------..............------------.(((((.......)))-))..(((((((((.........))))))..-..(((((((((((..(((.(((((.................)))))))).)))))))))))..)))..-- ( -24.53, z-score = -0.73, R) >droSec1.super_1 8459833 128 - 14215200 ---------------UUUUCCUCUAUAUA------------UGCAUGCACAUUUUAU-GCCCGCCGUCGUUCGUGCUGAAAAUGACUG-CUCGAAAUCAAUUUCAUGCGAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGUUGGCCG-- ---------------..............------------.(((((.......)))-))..(((((((((.........))))))..-..(((((((((((..(((.(((((.................)))))))).)))))))))))..)))..-- ( -24.53, z-score = -0.73, R) >droYak2.chr2R 10884246 140 - 21139217 ---------------UUUUCCUCUAUAUAUGUAUGUAUGUGUGCAUGCACAUUUUAU-GCCCGCCAUCGCUCGUGCUGAAAAUUACUG-CUCGAAAUCAAUUUCAUGCGAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGUUGGCCG-- ---------------...............(((((.((((((....))))))..)))-))..((((..((..(((.(....).))).)-).(((((((((((..(((.(((((.................)))))))).))))))))))).))))..-- ( -29.73, z-score = -1.48, R) >droEre2.scaffold_4845 7679274 132 + 22589142 ---------------UUUUCCUCUAUAUAUGUA--------UGCAUGCACAUUUCAU-GCCCGCCAUCGUUCGUGCUGAAAAUUACUG-CUCGAAAUCAAUUUCAUGCGAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGUUGGCCG-- ---------------..................--------.(((((.......)))-))..((((.(....(.((...........)-).)((((((((((..(((.(((((.................)))))))).))))))))))).))))..-- ( -23.73, z-score = -0.60, R) >droAna3.scaffold_13266 17500731 154 + 19884421 UUUGCUUUCUAUAUAUAUUUUUGUAUAUAUAGUU-UAAAUGGGUAUGGAUAUUUUAU-CCCUGGCAUCGCUCGUGCUGAAAAUUACUG-CUCGAAAUCAAUUUCAUGCGAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGUUGGCCA-- ........((((((((((....))))))))))..-.....(((.(((((...)))))-))).(((...((..(((.(....).))).)-).(((((((((((..(((.(((((.................)))))))).)))))))))))...))).-- ( -32.83, z-score = -1.62, R) >dp4.chr3 8837313 116 - 19779522 ---------------------------------------UACCCCUAUAUAUAUUCG-CAUAUAUAUUGCUCGUGCUGCAAAUUUCUG-CUCGAAAUCAAUUUCUUGCGAUUGAAGCACUUAAUUUGUACCAAUUCAUUAAUUGAUUUCGUUCGCCA-- ---------------------------------------......(((((((.....-.)))))))((((.......))))......(-(.(((((((((((...((.(((((..(((.......)))..)))))))..)))))))))))...))..-- ( -22.70, z-score = -2.45, R) >droPer1.super_4 2722319 116 - 7162766 ---------------------------------------UACCCCUAUAUAUAUUCG-CAUAUAUAUUGCUCGUGCUGCAAAUUUCUG-CUCGAAAUCAAUUUCUUGCGAUUGAAGCACUUAAUUUGUACCAAUUCAUUAAUUGAUUUCGUUCUCCA-- ---------------------------------------......(((((((.....-.)))))))((((.......))))......(-..(((((((((((...((.(((((..(((.......)))..)))))))..)))))))))))..)....-- ( -21.40, z-score = -2.69, R) >droWil1.scaffold_180699 2369212 134 - 2593675 ---------------UUUCUCUCUGUAGAUA--------UCCAUAUAUCUGUGGUGUGUAUGUUUGCCUCUCGAGCUGAAAAUUACUGCCUCGAAAUCAAUUUCAUGCAAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGUUGCCUG-- ---------------((((.(((.(((((((--------(.(((((.......))))).)))))))).....)))..))))......((..(((((((((((..(((.(((((.................)))))))).)))))))))))..))...-- ( -29.03, z-score = -2.87, R) >droVir3.scaffold_12875 9818769 114 - 20611582 -------------------------------------------UUUAUAUAUUUUUU-CCUGACCGGCGCUCAAGCUGAAAAUUAUUA-CUCGAAAUCAAUUUCAUACAAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGCUGCCGCUG -------------------------------------------..............-......((((((....))............-..(((((((((((......(((((.................)))))....)))))))))))..))))... ( -18.73, z-score = -1.90, R) >droMoj3.scaffold_6496 9636982 114 + 26866924 -------------------------------------------UUUAUAUAUUUUUU-CCUGGUCGGCGCUCAUGCUGAAAAUUAUUA-CUCGAAAUCAAUUUCGUACAAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCACUGCCGCUG -------------------------------------------..............-...((.((((((....))............-...((((((((((......(((((.................)))))....))))))))))...)))))). ( -19.53, z-score = -1.87, R) >droGri2.scaffold_15245 3024299 120 - 18325388 -------------------------------------UUUAUACAUAUAUAUAUUUU-CCUGGCCGUCGCUCGAACUGAAAAUUAUUA-CUCGAAAUCAAUUUCGUACAAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUUGCUGCCACCA -------------------------------------....................-..((((((.....))...............-..(((((((((((......(((((.................)))))....)))))))))))..))))... ( -13.53, z-score = -0.21, R) >consensus _______________UUUU_CUCUAUAUA____________UGCAUAUAUAUUUUAU_CCCCGCCAUCGCUCGUGCUGAAAAUUACUG_CUCGAAAUCAAUUUCAUGCGAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGUUGGCCG__ ...........................................................................................(((((((((((......(((((.................)))))....)))))))))))......... (-11.00 = -10.79 + -0.21)
| Location | 10,950,211 – 10,950,308 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 95.38 |
| Shannon entropy | 0.06364 |
| G+C content | 0.48824 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -28.87 |
| Energy contribution | -29.20 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
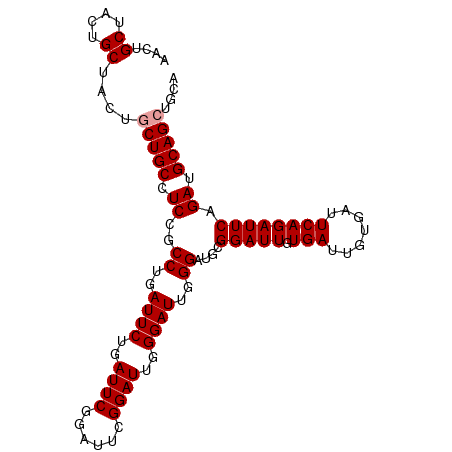
>dm3.chr2R 10950211 97 - 21146708 AACUGCUACUGCU----CUGCCUCUGCCUGAUUCUGAUUCGGAUUGGGAUUGGGAUUGGGAUGCGGAUUGUGAUUGUGAUUCAGAUUCAGAUGCAGCUGCA ....((..(((((----(((..(((.((..((((..((((......))))..))))..))....((((..(....)..)))))))..)))).))))..)). ( -35.20, z-score = -2.77, R) >droSec1.super_1 8459961 101 - 14215200 AACUGCUACUGCUACUGCUGCCUCCGCCUGAUUCUGAUUCGGAUUCGGAUUGGGAUUGGGAUGCGGAUUGUGAUUGUGAUUCAGAUUCAGAUGCAGCUGAA ....((....))....(((((.((..((..((((..(((((....)))))..))))..))....(((((.(((.......)))))))).)).))))).... ( -32.00, z-score = -1.80, R) >droSim1.chr2R 9702530 101 - 19596830 AACUGCUACUGCUACUGCUGCCUCCGCCUGAUUCUGAUUCGGAUUCGGAUUGGGAUUGGGAUGCGGAUUGUGAUUGUGAUUCAGAUUCAGAUGCAGCUGCA ....((..((((..(((..((.....((..((((..(((((....)))))..))))..))..))((((..(....)..)))).....)))..))))..)). ( -33.20, z-score = -1.68, R) >consensus AACUGCUACUGCUACUGCUGCCUCCGCCUGAUUCUGAUUCGGAUUCGGAUUGGGAUUGGGAUGCGGAUUGUGAUUGUGAUUCAGAUUCAGAUGCAGCUGCA ...(((..((((.....(((..((..((..((((..((((......))))..))))..))....((((..(....)..)))).))..)))..))))..))) (-28.87 = -29.20 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:07 2011