| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,892,673 – 2,892,779 |
| Length | 106 |
| Max. P | 0.990317 |

| Location | 2,892,673 – 2,892,779 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 67.52 |
| Shannon entropy | 0.57905 |
| G+C content | 0.57668 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.64 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990317 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
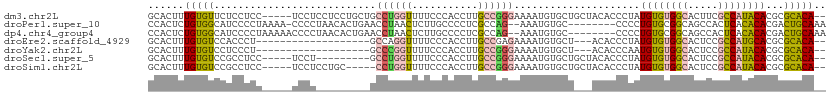
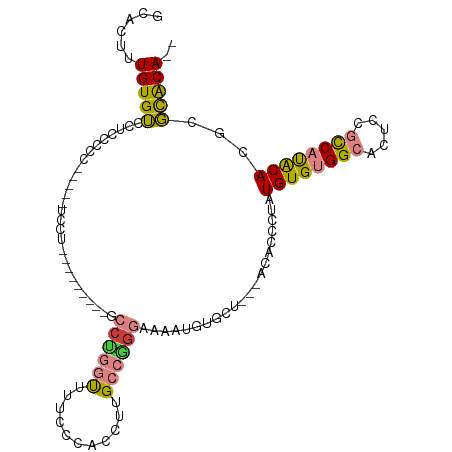
>dm3.chr2L 2892673 106 + 23011544 GCACUUUGUGUUCUCCUCC-----UCCUCCUCCUGCUGCCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCUGCUACACCCUAUGUGUGGCACUUCGCCAUACACGCGCACA-- ......(((((.(......-----..........((.((...((((((((........))))))))..)).)).........((((((((.....)))))))).).)))))-- ( -29.70, z-score = -2.81, R) >droPer1.super_10 98328 102 - 3432795 CCACUCUGUGGCAUCCCCUAAAA-CCCCUAACACUGAACCUAACUCUUGCCCCUCGCCAG--AAAUGUGC--------CCCCUGUGCGGCAGCCACUCACACACGACUGCAAA .......(((((...........-....................(((.((.....)).))--)....(((--------(........)))))))))................. ( -16.60, z-score = -0.51, R) >dp4.chr4_group4 2292659 103 - 6586962 CCACUCUGUGGCAUCCCCUAAAAACCCCUAACACUGAACCUAACUCUUGCCCCUCGCCAG--AAAUGUGC--------CCCCUGUGCGGCAGCCACUCACACACGACUGCAAA .......(((((................................(((.((.....)).))--)....(((--------(........)))))))))................. ( -16.60, z-score = -0.60, R) >droEre2.scaffold_4929 2940002 89 + 26641161 GCACUUUGUGUCCACCCU-------------------GCCAGGUUUUCCCACCUUGCCGAGAAAAUGUGCU---ACACCCUAUGUGUGGCACUCCGCCAUGCACGCGCACA-- ......(((((.(...(.-------------------((.((((......)))).)).).......(((((---((((.....)))))))))............).)))))-- ( -24.20, z-score = -0.94, R) >droYak2.chr2L 2887784 89 + 22324452 GCACUUUGUGUCCUCCCU-------------------GCCCGGUUUUCCCACCUUGCCGGGAAAAUGUGCU---ACACCCAAUGUGUGGCACUCCGCCAUACACGCGCACA-- ((.(..(((((..((((.-------------------((..(((......)))..)).))))....(((((---((((.....)))))))))......))))).).))...-- ( -29.40, z-score = -2.55, R) >droSec1.super_5 1051460 97 + 5866729 GCACUUUGUGUCCGCCUCC-----UCCU---------GCCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCUGCUACACCCUAUGUGUGGCACUCCGCCAUACACGCGCACA-- ......(((((.((.....-----....---------((...((((((((........))))))))..))............((((((((.....)))))))))).)))))-- ( -28.50, z-score = -1.64, R) >droSim1.chr2L 2853587 101 + 22036055 GCACUUUGUGUCCGCCUCC-----UCCUCCUGC-----CCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCUGCUACACCCUAUGUGUGGCACUCCGCCAUACACGCGCACA-- ......(((((.((.....-----...(((((.-----(..(((......)))..).)))))..(((.((((((((((.....))))))))....)))))...)).)))))-- ( -27.90, z-score = -1.71, R) >consensus GCACUUUGUGUCCUCCCCC_____UCCU_________GCCUGGUUUUCCCACCUUGCCGGGAAAAUGUGCU___ACACCCUAUGUGUGGCACUCCGCCAUACACGCGCACA__ ......(((((...........................((((((...........)))))).....................((((((((.....))))))))...))))).. (-16.74 = -16.64 + -0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:32 2011