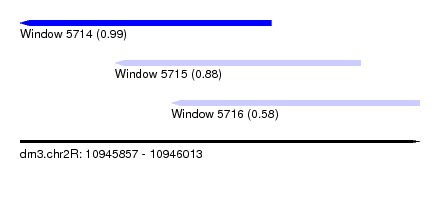
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,945,857 – 10,946,013 |
| Length | 156 |
| Max. P | 0.986847 |

| Location | 10,945,857 – 10,945,955 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 58.82 |
| Shannon entropy | 0.82112 |
| G+C content | 0.46335 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -7.36 |
| Energy contribution | -9.25 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
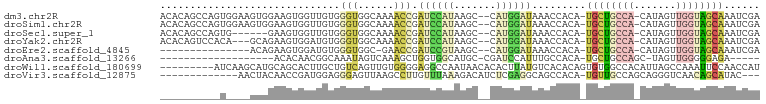
>dm3.chr2R 10945857 98 - 21146708 CAAAACCGAUCCAUAAGCC-AUGGAUA-AACCACAUGCUGCCACAUAGU-UGGUAGCA--AAUCGAGUUUACGAGGAAACUGUUGCUCAGAUGUGGCAAAAAU- ........((((((.....-)))))).-..(((((((((((((......-))))))).--....((((..((.((....)))).))))..)))))).......- ( -32.60, z-score = -4.34, R) >droSim1.chr2R 9698217 97 - 19596830 CAAAACCGAUCCAUAAGCC-AUGGAUA-AACCACAUGCUGCCACAUAGU-UGGUAGCA--AAUCGAGUUUACGAGGAAACUGUUGCUCAGAUGUGGCAAAAU-- ........((((((.....-)))))).-..(((((((((((((......-))))))).--....((((..((.((....)))).))))..))))))......-- ( -32.60, z-score = -4.27, R) >droSec1.super_1 8455667 97 - 14215200 CAAAACCGAUCCAUAAGCC-AUGGAUA-AACCACAUGCUGCCACAUAGU-UGGUAGCA--AAUCGAGUUUACGAGGAAACUGUUGCUCAGAUAUGGCAAAAU-- ................(((-(((....-.......((((((((......-))))))))--....((((..((.((....)))).))))...)))))).....-- ( -29.40, z-score = -3.65, R) >droYak2.chr2R 10879454 97 - 21139217 CAAAACCGAUCCAUAAGCC-AUGGAUA-AACCACAUGCUGCCACAUAGU-UGGUAGCA--AAUCGAGUUUAUGCGAAAACUGUUGCUCAGAUGUGGCAAAAU-- ........((((((.....-)))))).-..(((((((((((((......-))))))).--....((((....(((.....))).))))..))))))......-- ( -28.40, z-score = -2.94, R) >droEre2.scaffold_4845 7675327 95 + 22589142 -CGAACCGAUCCGUAAGCC-AUGGAUA-AACCACAUGCUGCCACAUAGU-UGGUAGCA--AAUCGAGUUUAUGCGGAAACUGUUGCUCAGAUGUGGCAAAU--- -.......((((((.....-)))))).-..(((((((((((((......-))))))).--....((((....(((....).)).))))..)))))).....--- ( -30.10, z-score = -2.55, R) >droAna3.scaffold_13266 17495885 78 + 19884421 -------------CAUGCCGAUCCAUU-UGCCACAUGCUGCCAGCUAGU-UGGGGG-------AGAGCUCCCAUAGCAGCAGCAACUUGG----GGCGAAGACU -------------............((-((((.((((((((..((((..-((((((-------....)))))))))).))))))...)).----)))))).... ( -30.40, z-score = -1.47, R) >droWil1.scaffold_180699 2363259 87 - 2593675 -------------UGGGGAGGCCAAUA-ACACACUUA-UGUCACACAGUGUGGCCACAUUAGCCAAAUUCCAACCAUGUCUACAGCCGAAAU--GGCCUAAAUU -------------((((..(((.(((.-.((((((..-........)))))).....))).))).........((((.((.......)).))--)))))).... ( -17.70, z-score = 0.64, R) >droVir3.scaffold_12875 9812646 80 - 20611582 ---------UUUAAAGACAUCUCGAGGCAGCCACAUGUUGCCAGCAGGGUCAACAGCAUACCACACGCUAACAAACAGAACAGAGCUGC--------------- ---------........((.(((..((((((.....))))))(((..(((.........)))....))).............))).)).--------------- ( -15.90, z-score = -0.42, R) >consensus _AAAACCGAUCCAUAAGCC_AUGGAUA_AACCACAUGCUGCCACAUAGU_UGGUAGCA__AAUCGAGUUUACGAGGAAACUGUUGCUCAGAUGUGGCAAAAA__ ................(((................((((((((.......))))))))......((((................))))......)))....... ( -7.36 = -9.25 + 1.89)
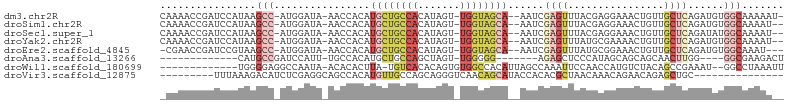
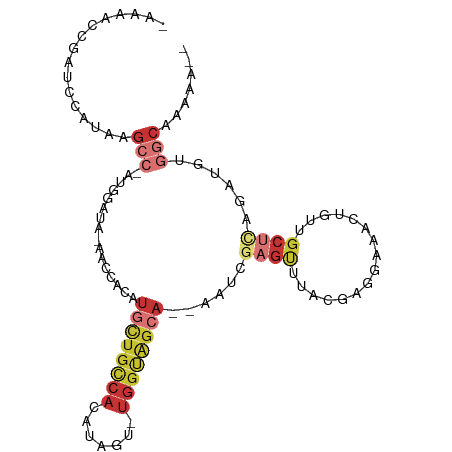
| Location | 10,945,894 – 10,945,990 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 55.86 |
| Shannon entropy | 0.87959 |
| G+C content | 0.49408 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -7.43 |
| Energy contribution | -8.34 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.94 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
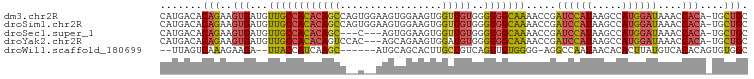
>dm3.chr2R 10945894 96 - 21146708 ACACAGCCAGUGGAAGUGGAAGUGGUUGUGGGUGGCAAAACCGAUCCAUAAGC--CAUGGAUAAACCACA-UGCUGCCA-CAUAGUUGGUAGCAAAUCGA .((((((((.(.........).)))))))).((((........((((((....--.))))))...)))).-((((((((-......))))))))...... ( -33.30, z-score = -2.10, R) >droSim1.chr2R 9698253 96 - 19596830 ACACAGCCAGUGGAAGUGGAAGUGGUUGUGGGUGGCAAAACCGAUCCAUAAGC--CAUGGAUAAACCACA-UGCUGCCA-CAUAGUUGGUAGCAAAUCGA .((((((((.(.........).)))))))).((((........((((((....--.))))))...)))).-((((((((-......))))))))...... ( -33.30, z-score = -2.10, R) >droSec1.super_1 8455703 90 - 14215200 ACACAGCCAGUG------GAAGUGGUUGUGGGUGGCAAAACCGAUCCAUAAGC--CAUGGAUAAACCACA-UGCUGCCA-CAUAGUUGGUAGCAAAUCGA .((((((((...------....)))))))).((((........((((((....--.))))))...)))).-((((((((-......))))))))...... ( -32.20, z-score = -2.36, R) >droYak2.chr2R 10879490 93 - 21139217 ACACAGUCCACA---GCAGAAGUGGAUGUGGGUGGCAAAACCGAUCCAUAAGC--CAUGGAUAAACCACA-UGCUGCCA-CAUAGUUGGUAGCAAAUCGA .((((.(((((.---......))))))))).((((........((((((....--.))))))...)))).-((((((((-......))))))))...... ( -32.30, z-score = -2.91, R) >droEre2.scaffold_4845 7675362 80 + 22589142 ---------------ACAGAAGUGGAUGUGGGUGGC-GAACCGAUCCGUAAGC--CAUGGAUAAACCACA-UGCUGCCA-CAUAGUUGGUAGCAAAUCGA ---------------...((.((((.....(((...-..))).((((((....--.))))))...)))).-((((((((-......))))))))..)).. ( -26.60, z-score = -2.02, R) >droAna3.scaffold_13266 17495919 73 + 19884421 -------------------ACACAACGGCAAAUAGUCAAAGCUGGUGGCAUGC-CGAUCCAUUUGCCACA-UGCUGCCAGC-UAGUUGGGGGAGA----- -------------------...(((((((.....)))..((((((..(((((.-(((.....)))...))-)))..)))))-).)))).......----- ( -28.30, z-score = -2.02, R) >droWil1.scaffold_180699 2363284 91 - 2593675 ---------AUCAAGCAUGCAGCACUUGCUGUCAGUUGUGGGGAGGCCAAUAACACACUUAUGUCACACAGUGUGGCCACAUUAGCCAAAUUCCAACCAU ---------....(((..(((((....)))))..)))((((((((((.(((..((((((..........)))))).....))).)))....)))..)))) ( -21.50, z-score = 0.64, R) >droVir3.scaffold_12875 9812674 83 - 20611582 -------------AACUACAACCGAUGGAGGGAGUUAAGCCUUGUUUAAAGACAUCUCGAGGCAGCCACA-UGUUGCCAGCAGGGUCAACAGCAUAC--- -------------((((....((......)).))))..((((((((....((....))..((((((....-.))))))))))))))...........--- ( -20.50, z-score = -0.58, R) >consensus __________U____GUAGAAGUGGUUGUGGGUGGCAAAACCGAUCCAUAAGC__CAUGGAUAAACCACA_UGCUGCCA_CAUAGUUGGUAGCAAAUCGA ..............................(((......))).((((((.......)))))).........((((((((.......))))))))...... ( -7.43 = -8.34 + 0.91)
| Location | 10,945,916 – 10,946,013 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 74.38 |
| Shannon entropy | 0.45152 |
| G+C content | 0.49511 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.78 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
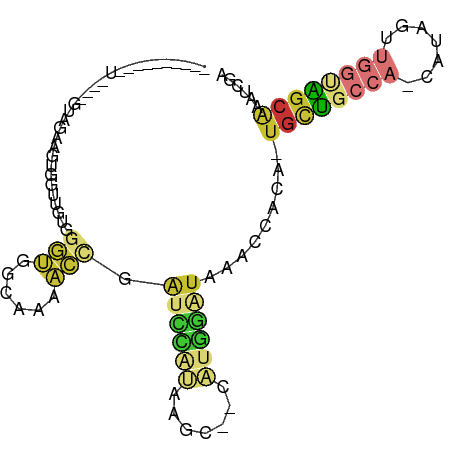
>dm3.chr2R 10945916 97 - 21146708 CAUGACACAGAAGUGAUGUUGCCACACAGCCAGUGGAAGUGGAAGUGGUUGUGGGUGGCAAAACCGAUCCAUAAGCCAUGGAUAAACCACA-UGCUGC .......(((..(((.(((..((.((((((((.(.........).))))))))))..)))......((((((.....))))))....))).-..))). ( -32.50, z-score = -1.74, R) >droSim1.chr2R 9698275 97 - 19596830 CAUGACACAGAAGUGAUGUUGCCACACAGCCAGUGGAAGUGGAAGUGGUUGUGGGUGGCAAAACCGAUCCAUAAGCCAUGGAUAAACCACA-UGCUGC .......(((..(((.(((..((.((((((((.(.........).))))))))))..)))......((((((.....))))))....))).-..))). ( -32.50, z-score = -1.74, R) >droSec1.super_1 8455725 91 - 14215200 CAUGACACAGAAGUGAUGUUGCCACACAGC---C---AGUGGAAGUGGUUGUGGGUGGCAAAACCGAUCCAUAAGCCAUGGAUAAACCACA-UGCUGC .......(((..(((.(((..((.((((((---(---(.......))))))))))..)))......((((((.....))))))....))).-..))). ( -31.40, z-score = -1.88, R) >droYak2.chr2R 10879512 94 - 21139217 CAUGACACAGAAGUGAUGUUGCCACACAGUCCAC---AGCAGAAGUGGAUGUGGGUGGCAAAACCGAUCCAUAAGCCAUGGAUAAACCACA-UGCUGC .......(((..(((.(((..((.((((.(((((---.......)))))))))))..)))......((((((.....))))))....))).-..))). ( -31.50, z-score = -2.20, R) >droWil1.scaffold_180699 2363307 87 - 2593675 --UUAGUCAAAGAAGA--UUACCAUCAAGC------AUGCAGCACUUGCUGUCAGUUGUGGGG-AGGCCAAUAACACACUUAUGUCACACAGUGUGGC --.(((((......))--)))((((..(((------..(((((....)))))..)))))))..-..........((((((..........)))))).. ( -20.00, z-score = 0.60, R) >consensus CAUGACACAGAAGUGAUGUUGCCACACAGCC__C__AAGUGGAAGUGGUUGUGGGUGGCAAAACCGAUCCAUAAGCCAUGGAUAAACCACA_UGCUGC .......(((..(((...(((((((((((((...............))))))..))))))).....((((((.....))))))....)))....))). (-16.26 = -16.78 + 0.52)
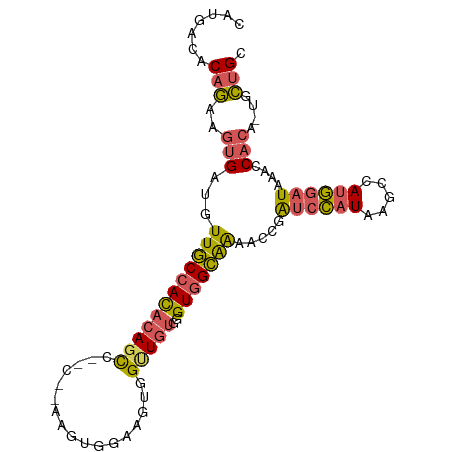
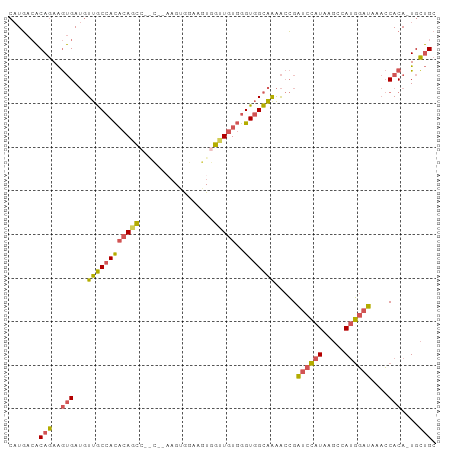
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:03 2011