| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,941,999 – 10,942,109 |
| Length | 110 |
| Max. P | 0.981254 |

| Location | 10,941,999 – 10,942,109 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.68 |
| Shannon entropy | 0.52981 |
| G+C content | 0.45182 |
| Mean single sequence MFE | -33.99 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
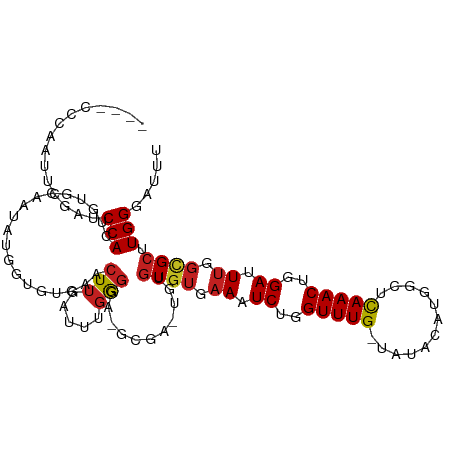
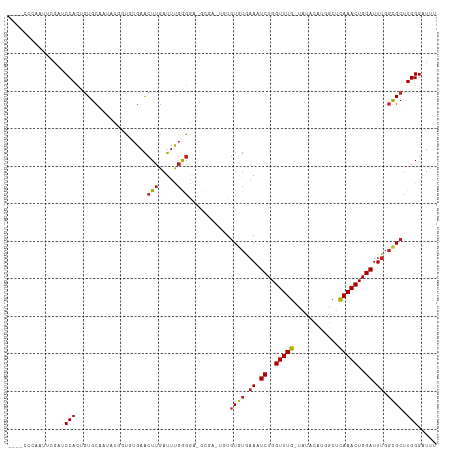
>dm3.chr2R 10941999 110 + 21146708 -UG-CCCAAUUCGAUCCACUGUGCAAUAUCUUGUGAACUUGAUUUGGGGA-GCGAAUGUGUGUGAAAUCUGGUUUG-UGUACAUGACUCAAACUGGAUUUGGCGCUUGGGAUUU -..-(((((((((.(((.(((..(((..((....))..)))...))))))-.)))))..((((.(((((..(((((-.((.....)).)))))..))))).)))).)))).... ( -37.70, z-score = -3.66, R) >droPer1.super_4 2716011 108 + 7162766 --UACCUGAUCUGUACCACUGUGCAGCUCGACUUGAACUUGAUUCGGGGGAGCGA--GUGUGUGAAAUCUGGUUUG-UAUACAUGGCUCAAACUGGAUUUGGCGCUUGGGAUU- --..((..((((((((....)))))((((..((((((.....)))))).))))))--..((((.(((((..(((((-...........)))))..))))).)))))..))...- ( -36.60, z-score = -2.75, R) >dp4.chr3 8830944 108 + 19779522 --UACCUGAUCUGUACCACUGUGCAGCUCGACUUGAACUUGAUUCGGGGGAGCGA--GUGUGUGAAAUCUGGUUUG-UAUACAUGGCUCAAACUGGAUUUGGCGCUUGGGAUU- --..((..((((((((....)))))((((..((((((.....)))))).))))))--..((((.(((((..(((((-...........)))))..))))).)))))..))...- ( -36.60, z-score = -2.75, R) >droAna3.scaffold_13266 17493433 113 - 19884421 CCGAUUGUUUUGGUUCCACUGUGCCAUAAAGUCUGAACUUGAUUUGGGGAAGCGAAUGUGUGUGAAAUCUGGUUUG-UAUACAUGGCUCAAACUGGAUUUGGCGCUUGGGAUUU ((((.....))))(((((..((((((..((((....))))((((..(.((.((..(((((((..((......))..-))))))).))))...)..)))))))))).)))))... ( -33.30, z-score = -2.03, R) >droEre2.scaffold_4845 7671152 109 - 22589142 ---CGCCCAUUCGAUCCACCGUGCAAUAUCUUGUGAACUUGAUUUGGGGA-GCGAAUGUGUGUGAAAUCUGGUUUG-UGUACAUGGCUCAAACUGGAUUUGGCGCUUGGGAUUU ---..((((((((.(((.(((..(((..((....))..)))...))))))-.)))))..((((.(((((..(((((-.((.....)).)))))..))))).))))..))).... ( -40.00, z-score = -3.50, R) >droYak2.chr2R 10874864 110 + 21139217 -UG-CCCAAUUCGAUCCACUGUGCAAUAUCUUGUGAACUUGAUUUGGGGA-GCGAAUGUGUGUGAAAUCUGGUUUG-UGUACAUGGCUCAAACUGGAUUUGGCGCUUGGGAUUU -..-(((((((((.(((.(((..(((..((....))..)))...))))))-.)))))..((((.(((((..(((((-.((.....)).)))))..))))).)))).)))).... ( -38.00, z-score = -3.40, R) >droSec1.super_1 8451645 112 + 14215200 UUGUCCCAAUUCGAUCCACUGUGCAAUAUCUUGUGAACUUGAUUUGGGGA-GCGAAUGUGUGUGAAAUCUGGUUUG-UGUACAUGGCUCAAACUGGAUUUGGCGCUUGGGAUUU ..(((((((((((.(((.(((..(((..((....))..)))...))))))-.)))))..((((.(((((..(((((-.((.....)).)))))..))))).)))).)))))).. ( -40.30, z-score = -4.11, R) >droSim1.chr2R 9694662 112 + 19596830 UUGUCCCAAUUCGAUCCACUGUGCAAUAUCUUGUGAACUUGAUUUGGGGA-GCGAAUGUGUGUGAAAUCUGGUUUG-UGUACAUGGCUCAAACUGGAUUUGGCGCUUGGGAUUU ..(((((((((((.(((.(((..(((..((....))..)))...))))))-.)))))..((((.(((((..(((((-.((.....)).)))))..))))).)))).)))))).. ( -40.30, z-score = -4.11, R) >droVir3.scaffold_12875 9810130 92 + 20611582 -----------UGUACCACUGUGCU---UGGCCUGAACUUGAUUUGAGAC-------UUGUGUGAAUUCUGGUUUG-AAACCAUGUCUCAAACUGGAUUUGGCGCUUGGGAUUU -----------.((((....)))).---...((..(.(((.....)))..-------..((((.((.((..(((((-(.((...)).))))))..)).)).)))))..)).... ( -23.70, z-score = -1.80, R) >droMoj3.scaffold_6496 9626497 92 - 26866924 -----------UGCACCACGGUGCU---UGGCCUGAACUUGAUUUGAGAC-------UUGUGUGAAUUCUGGUUUG-AAGCCAUGGCCCAAACUGGAUUUGGCGCUUGGGAUUU -----------.((((....)))).---...((..(.(((.....)))..-------..((((.((.((..(((((-..((....)).)))))..)).)).)))))..)).... ( -29.00, z-score = -1.83, R) >droGri2.scaffold_15245 3017183 97 + 18325388 ------CACUUUGCUCCAAUGUGCA---UGGCCUGAACUUGAUUUGAGAC-------UUGUGUGAAUUCUGGUUUG-AAGCCGUGGCUCAAACUGGAUUUGGUGCUUGGGAUUU ------..((..((.((((...(((---..(.((.((......)).))..-------)..)))....((..(((((-(.((....))))))))..)).)))).))..))..... ( -26.40, z-score = -1.25, R) >droWil1.scaffold_180699 2359211 104 + 2593675 --------CCAGUGUCCAUAAUG-AUCUUGGUUUGAGCUUGAUUCCACGGUUUGC-UUUUUAAACAAUCUGGUUUGCUCGACAUGGCUUAAACUGGAUUUGGCGCUUGGGAUUU --------((((((.(((....(-((((.(((((((((((((..(((..(((((.-....)))))....))).....)))....))))))))))))))))))))).)))..... ( -26.00, z-score = -0.74, R) >consensus ____CCCAAUUCGAUCCACUGUGCAAUAUGGUGUGAACUUGAUUUGGGGA_GCGA_UGUGUGUGAAAUCUGGUUUG_UAUACAUGGCUCAAACUGGAUUUGGCGCUUGGGAUUU ...............(((...................(((.....)))...........((((.((.((..(((((............)))))..)).)).)))).)))..... (-13.57 = -13.70 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:24:00 2011