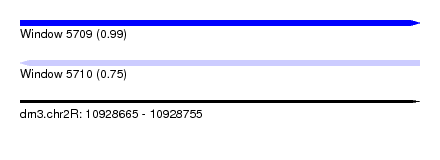
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,928,665 – 10,928,755 |
| Length | 90 |
| Max. P | 0.991359 |

| Location | 10,928,665 – 10,928,755 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 82.42 |
| Shannon entropy | 0.23956 |
| G+C content | 0.46740 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -23.82 |
| Energy contribution | -24.60 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
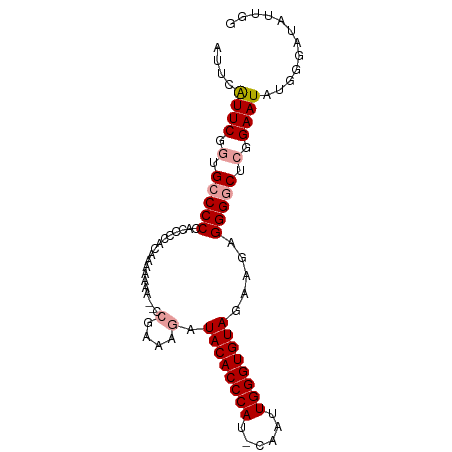
>dm3.chr2R 10928665 90 + 21146708 AUUCGUUCGCAGACCCCUCACCACAGAAAAA--CCGAAAGAUACACCCAUUCAAUUGGGUGUAGAAGAGGGGCACGGAAUAUGGAAUAUUGG .(((((...(...((((((............--.(....).((((((((......))))))))...))))))....)...)))))....... ( -29.20, z-score = -3.22, R) >droSim1.chr2R 9681382 89 + 19596830 AUUCAUUCGGUGCCCCAACCCCACAAAGAAA--CCGAAAGAUACACCCAU-CAAUUGGGUGUAGAAGAGGGGCUCGGAAUAUGGGAUAUUGG ....((((.(.(((((...............--.(....).((((((((.-....)))))))).....))))).).))))............ ( -30.80, z-score = -2.38, R) >droSec1.super_1 8438518 84 + 14215200 AUUCAUUCGGUGCCCCCAACCCACAAAAAAAAACAGAAAGAUACACCCAU-CAAUUGGGUGUAGAAGAGGGGCUCGGAAUAUUGG------- ....((((.(.((((((................(.....).((((((((.-....))))))))...).))))).).)))).....------- ( -26.80, z-score = -2.81, R) >consensus AUUCAUUCGGUGCCCCCACCCCACAAAAAAA__CCGAAAGAUACACCCAU_CAAUUGGGUGUAGAAGAGGGGCUCGGAAUAUGGGAUAUUGG ....((((.(.(((((..................(....).((((((((......)))))))).....))))).).))))............ (-23.82 = -24.60 + 0.78)

| Location | 10,928,665 – 10,928,755 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Shannon entropy | 0.23956 |
| G+C content | 0.46740 |
| Mean single sequence MFE | -28.31 |
| Consensus MFE | -19.95 |
| Energy contribution | -21.40 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.748696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
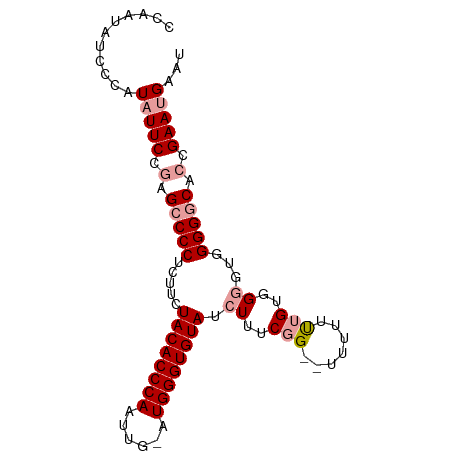
>dm3.chr2R 10928665 90 - 21146708 CCAAUAUUCCAUAUUCCGUGCCCCUCUUCUACACCCAAUUGAAUGGGUGUAUCUUUCGG--UUUUUCUGUGGUGAGGGGUCUGCGAACGAAU .............(((((.(((((((.((((((((((......)))))))).....(((--.....))).)).))))))).)).)))..... ( -28.40, z-score = -2.42, R) >droSim1.chr2R 9681382 89 - 19596830 CCAAUAUCCCAUAUUCCGAGCCCCUCUUCUACACCCAAUUG-AUGGGUGUAUCUUUCGG--UUUCUUUGUGGGGUUGGGGCACCGAAUGAAU ((...((((((((....(((((.......((((((((....-.))))))))......))--)))...))))))))..))............. ( -28.92, z-score = -1.49, R) >droSec1.super_1 8438518 84 - 14215200 -------CCAAUAUUCCGAGCCCCUCUUCUACACCCAAUUG-AUGGGUGUAUCUUUCUGUUUUUUUUUGUGGGUUGGGGGCACCGAAUGAAU -------....(((((.(.(((((.....((((((((....-.)))))))).(..((..(........)..))..)))))).).)))))... ( -27.60, z-score = -2.26, R) >consensus CCAAUAUCCCAUAUUCCGAGCCCCUCUUCUACACCCAAUUG_AUGGGUGUAUCUUUCGG__UUUUUUUGUGGGGUGGGGGCACCGAAUGAAU ...........(((((.(.(((((.....((((((((......)))))))).((..(((.......)))..))...))))).).)))))... (-19.95 = -21.40 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:59 2011