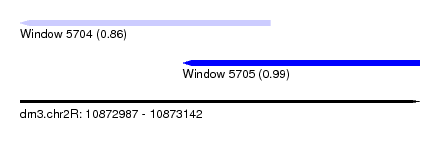
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,872,987 – 10,873,142 |
| Length | 155 |
| Max. P | 0.986386 |

| Location | 10,872,987 – 10,873,084 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.28 |
| Shannon entropy | 0.53439 |
| G+C content | 0.54406 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -10.95 |
| Energy contribution | -12.30 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10872987 97 - 21146708 -UAGGUCGACCUGACCUACAUCUGCUGGUGACAGCUUUGUCC----------------CCAGCUCCUGCCAGAACCACCCCACUUUCCUGGCCAGACUUGGGCAUAAUCGCAAU- -((((((.....)))))).....((((....)))).((((.(----------------((((.((..(((((...............)))))..)).))))).)))).......- ( -32.16, z-score = -2.76, R) >droPer1.super_2 3367619 91 + 9036312 CGGGGCUGACCUGACCUG-ACCUACAUACAUCCGCACUCUC-----------------UCUGUCUCUCUC----UUGUCCUGUUGGUGCGAC--AACAUUGCCAAAAUGGCAAUA ((((.....)))).....-.............((((((..(-----------------...(.(......----..).)..)..))))))..--...((((((.....)))))). ( -20.50, z-score = -0.87, R) >dp4.chr3 14310390 91 + 19779522 CGGGGCUGACCUGACCUG-ACCUACAUACAUCCGCAAUCUC-----------------UCACUCUCUCUC----UUGCCCUGUUGGUGCGAC--AACAUUGCCAAAAUGGCAAUA .(((((.((...((..((-(.....................-----------------)))...))..))----..)))))((((......)--)))((((((.....)))))). ( -19.00, z-score = -0.70, R) >droEre2.scaffold_4845 7608659 97 + 22589142 -UAGGUCGACCUGACCUACAUCUGCUGGUGACAGCUUUCUCU----------------CCUGCUCCUGCCAGAAGCACCCCACUUUUCUGGCCAGACUUUGGCAGAAUCGCAAA- -((((((.....)))))).....((((....)))).......----------------.(((((.((((((((((.........))))))).))).....))))).........- ( -30.80, z-score = -2.18, R) >droYak2.chr2R 10803499 113 - 21139217 -UAGGUCGACCUGACCUACAUCUGCUGGUGACAGCUUUCUCCCUUUCUCCCUUUCUCGCCUGGUCCUGCCAGAAGCACCCCACUUCUCUGGCCAGACUUUGGCAGAAUCGCAAA- -((((((.....)))))).....((((....)))).................((((.(((.((((..((((((.............))))))..))))..))))))).......- ( -36.62, z-score = -2.83, R) >droSec1.super_1 8385544 97 - 14215200 -UAGGUCGACCUGACCUACAUCUGCUGGUGACAGCUUUCUCC----------------CCAGCUCCUGCCGGAACCACCCCACUUUCCUGGCCAGACUUGGGUAGAAUCGCAAA- -((((((.....)))))).....((((....)))).((((.(----------------((((.((..(((((((..........)))).)))..)).))))).)))).......- ( -34.40, z-score = -3.02, R) >droSim1.chr2R 9633574 97 - 19596830 -UAGGUCGACCUGACCUAAAUCUGCUGGUGACAGCUUUCUCC----------------CCAGCUCCUGCCGGAACCACCCCACUUUCCUGGCCAGACUUGGGCAGAAUCGCAAA- -((((((.....)))))).....((((....)))).((((.(----------------((((.((..(((((((..........)))).)))..)).))))).)))).......- ( -34.70, z-score = -3.13, R) >consensus _UAGGUCGACCUGACCUACAUCUGCUGGUGACAGCUUUCUCC________________CCAGCUCCUGCCAGAACCACCCCACUUUUCUGGCCAGACUUUGGCAGAAUCGCAAA_ .((((((.....)))))).....((((....))))................................(((((...............)))))....................... (-10.95 = -12.30 + 1.35)

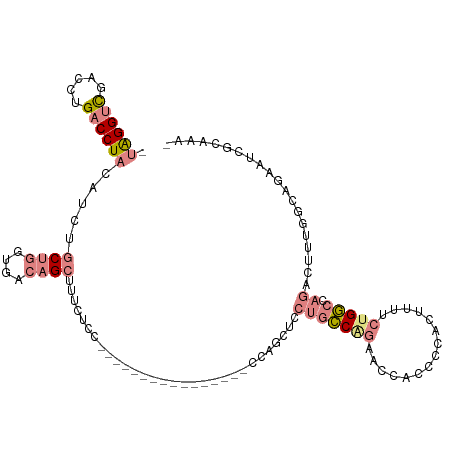
| Location | 10,873,050 – 10,873,142 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 94.98 |
| Shannon entropy | 0.08577 |
| G+C content | 0.60775 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -35.64 |
| Energy contribution | -35.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10873050 92 - 21146708 ---------GUUCCGCCAGUUCCGCAAUGACGCCAGCUGCCCUUUGUGCCGGUGGCGUGAUGCCGAAUAGGUCGACCUGACCUACAUCUGCUGGUGACAGC ---------....(((((((((.(((.(.((((((.(.((.......)).).)))))).)))).)).((((((.....)))))).....)))))))..... ( -35.60, z-score = -2.21, R) >droEre2.scaffold_4845 7608722 92 + 22589142 ---------GUUCCGCCAGUUCCGCAAUGACGCCAGCUGCCCUUUGUGCCGGUGGCGUGAUGCCGAAUAGGUCGACCUGACCUACAUCUGCUGGUGACAGC ---------....(((((((((.(((.(.((((((.(.((.......)).).)))))).)))).)).((((((.....)))))).....)))))))..... ( -35.60, z-score = -2.21, R) >droYak2.chr2R 10803578 101 - 21139217 GUUCCACCACUUCCGCCAGUUCCGCAAUGACGCCAGCUGCCCUUUGUGCCGGUGGCGUGAUGCCGAAUAGGUCGACCUGACCUACAUCUGCUGGUGACAGC .............(((((((((.(((.(.((((((.(.((.......)).).)))))).)))).)).((((((.....)))))).....)))))))..... ( -35.60, z-score = -1.77, R) >droSec1.super_1 8385607 92 - 14215200 ---------GUUCCGCCAGUUCCGCAAUGACGCCAGCUGCCCUUUGUGCCGGUGGCGUGAUGCCGAAUAGGUCGACCUGACCUACAUCUGCUGGUGACAGC ---------....(((((((((.(((.(.((((((.(.((.......)).).)))))).)))).)).((((((.....)))))).....)))))))..... ( -35.60, z-score = -2.21, R) >droSim1.chr2R 9633637 92 - 19596830 ---------GUUCCGCCAGUUCCGCAAUGACGCCAGCUGCCCUUUGUGCCGGUGGCGUGAUGCCGAGUAGGUCGACCUGACCUAAAUCUGCUGGUGACAGC ---------....(((((((((.(((.(.((((((.(.((.......)).).)))))).)))).)).((((((.....)))))).....)))))))..... ( -35.80, z-score = -2.30, R) >consensus _________GUUCCGCCAGUUCCGCAAUGACGCCAGCUGCCCUUUGUGCCGGUGGCGUGAUGCCGAAUAGGUCGACCUGACCUACAUCUGCUGGUGACAGC .............(((((((((.(((.(.((((((.(.((.......)).).)))))).)))).)).((((((.....)))))).....)))))))..... (-35.64 = -35.64 + 0.00)
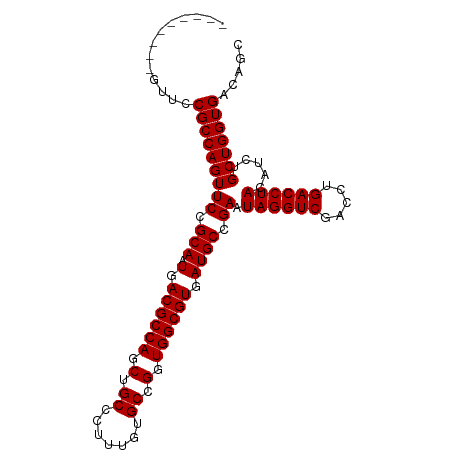
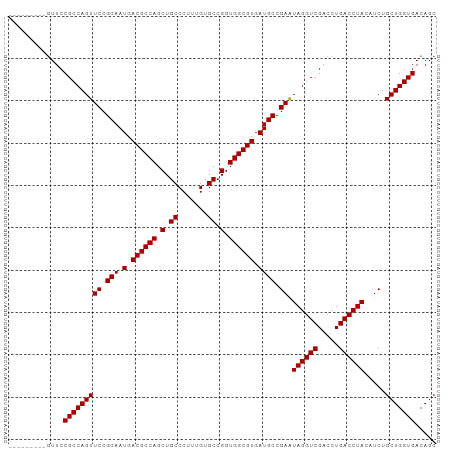
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:55 2011