| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,892,538 – 2,892,636 |
| Length | 98 |
| Max. P | 0.894843 |

| Location | 2,892,538 – 2,892,636 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Shannon entropy | 0.38301 |
| G+C content | 0.36315 |
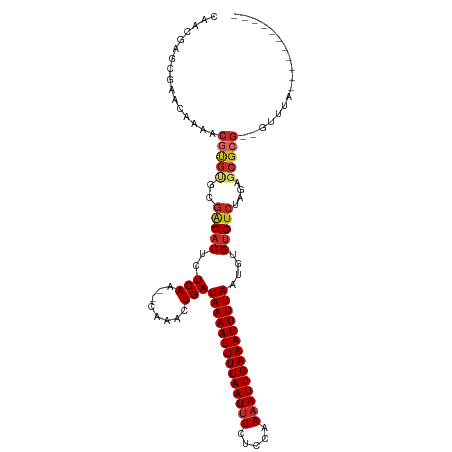
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.70 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
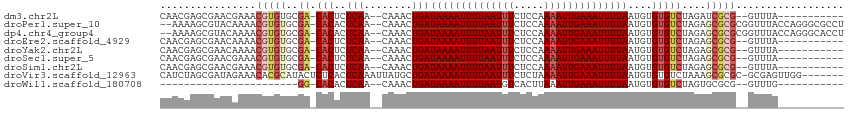
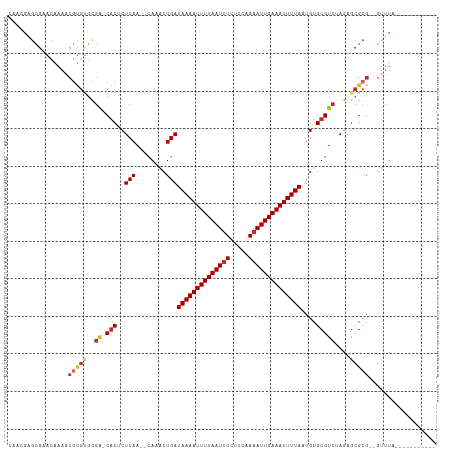
>dm3.chr2L 2892538 98 + 23011544 CAACGAGCGAACGAAACGUGUGCGA-CACUCUCAA--CAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAUCGCG--GUUUA----------- (..((((((.(((...))).)))((-(((.((((.--.....)))((((((((((((((.....))))))))))))))..).)))))....)))..--)....----------- ( -21.30, z-score = -1.54, R) >droPer1.super_10 98133 109 - 3432795 --AAAAGCGUACAAAACGUGUGCGA-CACACUCAA--CAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCGCGGUUUACCAGGGCGCCU --....(((..(.....((((((((-((((((((.--.....)))((((((((((((((.....))))))))))))))..)))))))....))))))((....)).)..))).. ( -35.00, z-score = -4.30, R) >dp4.chr4_group4 2292450 109 - 6586962 --AAAAGCGUACAAAACGUGUGCGA-CACACUCAA--CAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCGCGGUUUACCAGGGCACCU --....(((((((.....)))))((-((((((((.--.....)))((((((((((((((.....))))))))))))))..)))))))....))((.(((....)).).)).... ( -31.70, z-score = -3.60, R) >droEre2.scaffold_4929 2939877 98 + 26641161 CAACGAGCGAACAAAACGUGUGCGA-CACUCUCAA--CAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCG--GUUUA----------- ................(((((..((-(((.((((.--.....)))((((((((((((((.....))))))))))))))..).)))))....)))))--.....----------- ( -21.80, z-score = -1.74, R) >droYak2.chr2L 2887642 98 + 22324452 CAACGAGCGAACAAAACGUGUGCGA-CACUCUCAA--CAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCG--GUUUA----------- ................(((((..((-(((.((((.--.....)))((((((((((((((.....))))))))))))))..).)))))....)))))--.....----------- ( -21.80, z-score = -1.74, R) >droSec1.super_5 1051325 98 + 5866729 CAACGAGCGAACGAAACGUGUGCGA-CACUCUCAA--CAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCG--GUUUA----------- ................(((((..((-(((.((((.--.....)))((((((((((((((.....))))))))))))))..).)))))....)))))--.....----------- ( -21.80, z-score = -1.41, R) >droSim1.chr2L 2853452 98 + 22036055 CAACGAGCGAACGAAACGUGUGCGA-CACUCUCAA--CAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCG--GUUUA----------- ................(((((..((-(((.((((.--.....)))((((((((((((((.....))))))))))))))..).)))))....)))))--.....----------- ( -21.80, z-score = -1.41, R) >droVir3.scaffold_12963 19331834 106 - 20206255 CAUCUAGCGAUAGAAACACGCAUACUCUCACUCAAAUUAUGCUGAUAAAAUUUUAAUUUCUCUAAAAUUGAAAUUUUAAUGUGUGUCUAAAGCGCGC-GCGAGUUGG------- ......(((.((((.(((((((((.............)))))...((((((((((((((.....))))))))))))))..)))).))))...)))((-....))...------- ( -25.62, z-score = -2.69, R) >droWil1.scaffold_180708 7499312 75 - 12563649 -----------------------GG-CACACUCAA--CAAACUGAUAAAAUUUUAAUGUCACUUAAAUUGAAAUUUUAAUGUGUGUCUAGUGCGCG--GUUUG----------- -----------------------.(-(.((((..(--((.((...((((((((((((.........))))))))))))..)).)))..)))).)).--.....----------- ( -18.60, z-score = -2.93, R) >consensus CAACGAGCGAACAAAACGUGUGCGA_CACUCUCAA__CAAACUGAUAAAAUUUUAAUUUCUCCAAAAUUGAAAUUUUAAUGUGUGUCUAGAGCGCG__GUUUA___________ ......(((.((.....)).)))...............((((...((((((((((((((.....))))))))))))))..((((.......))))...))))............ (-12.46 = -12.70 + 0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:31 2011