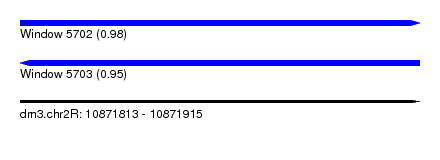
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,871,813 – 10,871,915 |
| Length | 102 |
| Max. P | 0.976595 |

| Location | 10,871,813 – 10,871,915 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.76 |
| Shannon entropy | 0.29044 |
| G+C content | 0.56027 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -31.55 |
| Energy contribution | -31.74 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
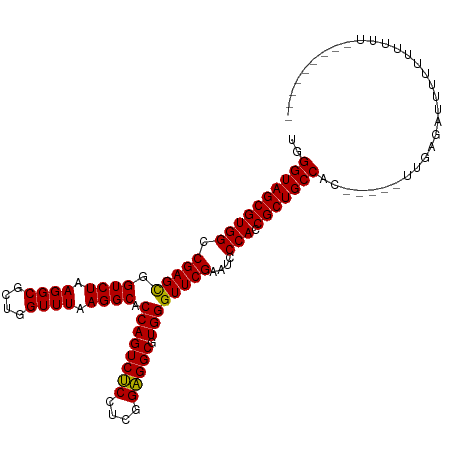
>dm3.chr2R 10871813 102 + 21146708 --------AAAAAAAAAAAUCUCAA-----GUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCCA --------.................-----((((((((((((((.......)))))...((..(((.(.(((((.......)))))).)))..)).))))..)))))........ ( -36.00, z-score = -2.42, R) >droSim1.chr2R 9632344 109 + 19596830 --AAAAAAAAAAAAAAAAAUCUCAA----AGUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCCA --.......................----.((((((((((((((.......)))))...((..(((.(.(((((.......)))))).)))..)).))))..)))))........ ( -36.00, z-score = -2.63, R) >droSec1.super_1 8384322 104 + 14215200 ------AAUAAAAAAAAAAUCCCAA-----GUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCCA ------...................-----((((((((((((((.......)))))...((..(((.(.(((((.......)))))).)))..)).))))..)))))........ ( -36.00, z-score = -2.50, R) >droYak2.chr2R 10802290 100 + 21139217 ----------AAAAAAAAAUCCUAA-----GUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCCA ----------...............-----((((((((((((((.......)))))...((..(((.(.(((((.......)))))).)))..)).))))..)))))........ ( -36.00, z-score = -2.41, R) >droEre2.scaffold_4845 7607465 100 - 22589142 ----------GAAAAAAAAUCGCA-----UGUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCCA ----------...........((.-----.((((((((((((((.......)))))...((..(((.(.(((((.......)))))).)))..)).))))..)))))))...... ( -36.20, z-score = -2.01, R) >dp4.chr3 3394036 105 + 19779522 ---------ACAAAAAGACACAUUGCUGUUCUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCC- ---------......(((.(((....))))))((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).)).- ( -33.50, z-score = -0.89, R) >droPer1.super_2 3366540 115 - 9036312 AAAAAAAAGGAAAAAAAGAUCAUUCUCUAUGUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCCU .......(((......(((....)))....((((((((((((((.......)))))...((..(((.(.(((((.......)))))).)))..)).))))..))))).....))) ( -37.70, z-score = -1.99, R) >droVir3.scaffold_12875 9737348 103 + 20611582 -----------AAAAAAAAUCCCAAUC-ACUUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCCC -----------................-....((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).)).. ( -32.40, z-score = -1.50, R) >droMoj3.scaffold_6496 9530814 100 - 26866924 -------AUAAAAAAA-AAUCUC-------UUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCAU -------.........-......-------.(((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).))). ( -34.00, z-score = -2.23, R) >droGri2.scaffold_15245 2945676 105 + 18325388 -------ACAAAAAAAGAAGCUCAA---AGUUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCACUCGGCCACGCUACCAA -------..................---..((((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).)))) ( -34.30, z-score = -1.74, R) >anoGam1.chr3L 27977688 104 + 41284009 ---------AAAAAGAAAAGUUUCAUUCGCAUGGCAGCGGUGGGAUUCGAACCCACGCCACCGAAGUGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACC-- ---------...................((.(((((((((((((.......)))......(..(.(((.(((((.......)))))))).)..)))))))..)))).))....-- ( -32.30, z-score = -1.10, R) >consensus _________AAAAAAAAAAUCUCAA_____GUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAGGAGACUGGUGCCUUAAACCAGCGCCUUAGACCGCUCGGCCACGCUACCCA ................................((.(((((((((.......)))))(((((..(((.(.(((((.......)))))).)))..))......)))..)))).)).. (-31.55 = -31.74 + 0.18)

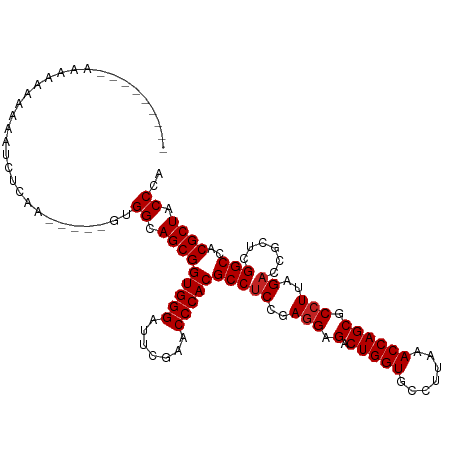
| Location | 10,871,813 – 10,871,915 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.76 |
| Shannon entropy | 0.29044 |
| G+C content | 0.56027 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -38.86 |
| Energy contribution | -38.61 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10871813 102 - 21146708 UGGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAC-----UUGAGAUUUUUUUUUUU-------- ..((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))..-----.................-------- ( -38.90, z-score = -1.27, R) >droSim1.chr2R 9632344 109 - 19596830 UGGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCACU----UUGAGAUUUUUUUUUUUUUUUUU-- ..((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))...----.......................-- ( -38.90, z-score = -1.38, R) >droSec1.super_1 8384322 104 - 14215200 UGGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAC-----UUGGGAUUUUUUUUUUAUU------ ..((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))..-----...................------ ( -38.90, z-score = -0.98, R) >droYak2.chr2R 10802290 100 - 21139217 UGGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAC-----UUAGGAUUUUUUUUU---------- ..((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))..-----...............---------- ( -38.90, z-score = -1.23, R) >droEre2.scaffold_4845 7607465 100 + 22589142 UGGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCACA-----UGCGAUUUUUUUUC---------- ..((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))...-----..............---------- ( -38.90, z-score = -1.16, R) >dp4.chr3 3394036 105 - 19779522 -GGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAGAACAGCAAUGUGUCUUUUUGU--------- -.((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))((((((....))).)))......--------- ( -41.70, z-score = -1.31, R) >droPer1.super_2 3366540 115 + 9036312 AGGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCACAUAGAGAAUGAUCUUUUUUUCCUUUUUUUU ..((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).))))))).....(((((........)))))......... ( -40.60, z-score = -1.15, R) >droVir3.scaffold_12875 9737348 103 - 20611582 GGGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAAGU-GAUUGGGAUUUUUUUU----------- ..((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))....-................----------- ( -38.90, z-score = -0.59, R) >droMoj3.scaffold_6496 9530814 100 + 26866924 AUGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAA-------GAGAUU-UUUUUUUAU------- .(((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))).-------......-.........------- ( -39.90, z-score = -2.09, R) >droGri2.scaffold_15245 2945676 105 - 18325388 UUGGUAGCGUGGCCGAGUGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAACU---UUGAGCUUCUUUUUUUGU------- ((((((((((..(((((..(.((..((.(((.......))).)))).)..)))))..))(((((.......))))).))))))))..---..................------- ( -40.10, z-score = -1.54, R) >anoGam1.chr3L 27977688 104 - 41284009 --GGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCACUUCGGUGGCGUGGGUUCGAAUCCCACCGCUGCCAUGCGAAUGAAACUUUUCUUUUU--------- --....(((((((..((((((....((.(((.......))).)).(((((....)))))..(((.......))))))))))))))))(((........))).....--------- ( -41.50, z-score = -2.07, R) >consensus UGGGUAGCGUGGCCGAGCGGUCUAAGGCGCUGGUUUAAGGCACCAGUCUCCUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAC_____UUGAGAUUUUUUUUUU_________ ..((((((((((.(((((.((((.((((....)))).)))).((((((((....))))).))))))))....))).)))))))................................ (-38.86 = -38.61 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:53 2011